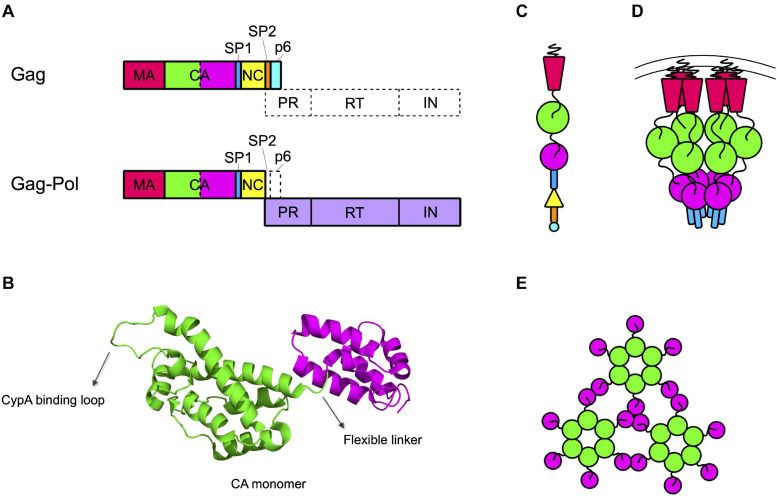
FIGURE 1.
Capsid forms throughout the HIV life cycle. (A) Gag and Gag-Pol precursors simplified structures. Gag precursor includes the matrix protein (MA), the capsid (CA, depicted with the NTD in green and the CTD in magenta), the spacer peptide 1 (SP1), the nucleocapsid (NC), the spacer peptide 2 (SP2), and the peptide 6 (p6). A frameshift during translation allows the production of Gag-Pol precursor, with a ratio of 1:20 with respect to the Gag precursor. In this structure the NC is fused to the protease (PR), the reverse transcriptase (RT), and the integrase (IN) domains. (B) Structure of CA monomer. CA is composed of two domains connected by a flexible linker: the NTD (in green), formed by a beta-hairpin and seven alpha-helices, and the CTD (in magenta), formed by four alpha-helices. The CypA binding loop in the NTD is indicated. PDB ID: 6WAP (Lu et al., 2020). (C) Schematic structure of the Gag precursor composed from top to bottom of MA, CA-NTD, CA-CTD, SP1, NC, SP2, and p6. (D) Schematic structure of a hexamer in the immature lattice, after the first proteolytic cleavage, which occurs between SP1 and NC. The MA are attached to the membrane through their myristoylated domain. Proceeding toward the center of the viral particle there are three hexameric structures composed by the CA-NTDs, CA-CTDs, and SP1. (E) Schematic top view of the mature capsid lattice where CA monomers are arranged in hexamers and are connected to each other through the NTDs, while the CTDs are involved in the interactions between hexamers.