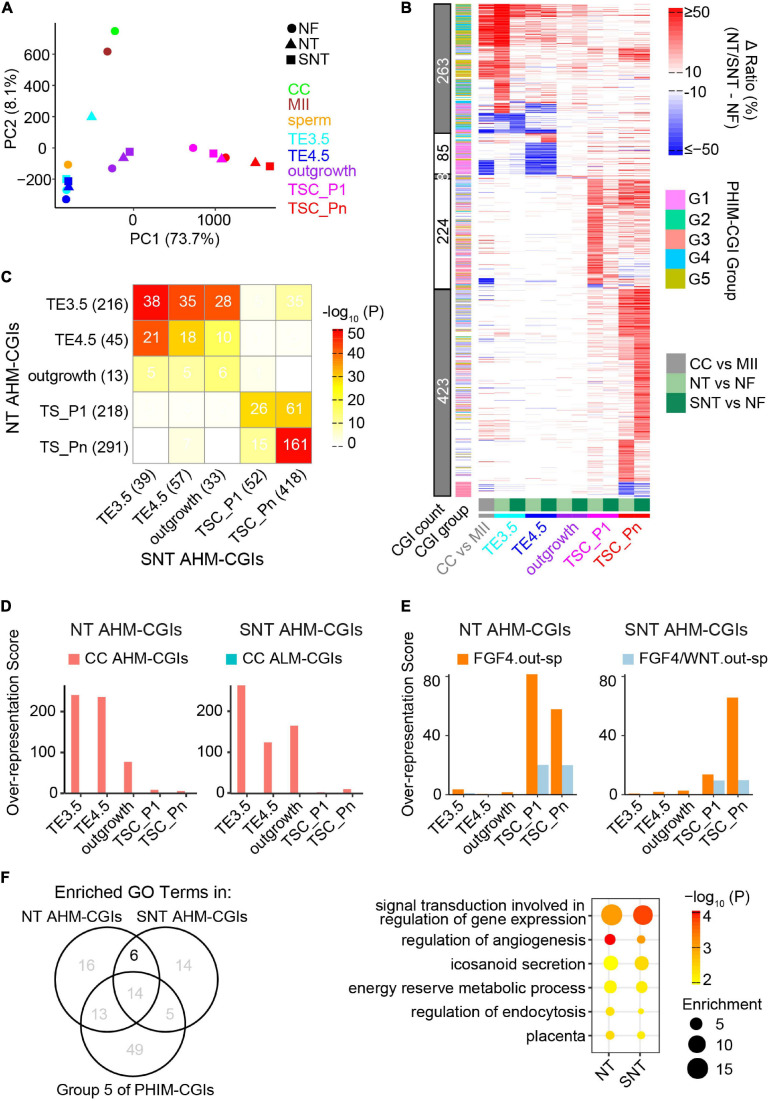
FIGURE 5.
Abnormal methylation in the donor genome is a barrier to methylation remodeling during NT TSC derivation. (A) PCA results of CGI methylation during NF, NT, and SNT TSC derivation. CC (cumulus cell), MII oocyte, and sperm are also included for comparison. Their methylation data are from GSE56697 (Wang et al., 2014). (B) A heat map showing methylation difference of aberrantly highly methylated (AHM)- and aberrantly lowly methylated (ALM)-CGIs (Definitions are in section “Results”). (C) A heat map showing the intersection between NT and SNT AHM-CGIs. Filled colors indicate the significance of the intersection (hypergeometric test). Numbers indicate CGI count. (D) CC AHM-CGIs are enriched in early stages (TE3.5, TE4.5, and outgrowth) of NT/SNT TSC derivation. (E) “FGF4.out-sp” CGIs are specifically enriched in AHM-CGIs of NT and SNT TSC_P1, and TSC_Pn. “FGF4.out-sp” denotes the CGIs that are highly methylated in ICM treated with FGF4 but not in EXE and EPI. (F) Venn diagram showing the intersection the GO terms for which NT/SNT AHM-CGIs and the Group 5 of PHIM-CGIs (Figure 4C) are significantly enriched. The numbers indicate GO term count. The bubble plot showing the six GO terms specifically common to the GO terms for which NT/SNT AHM-CGIs are significantly enriched.