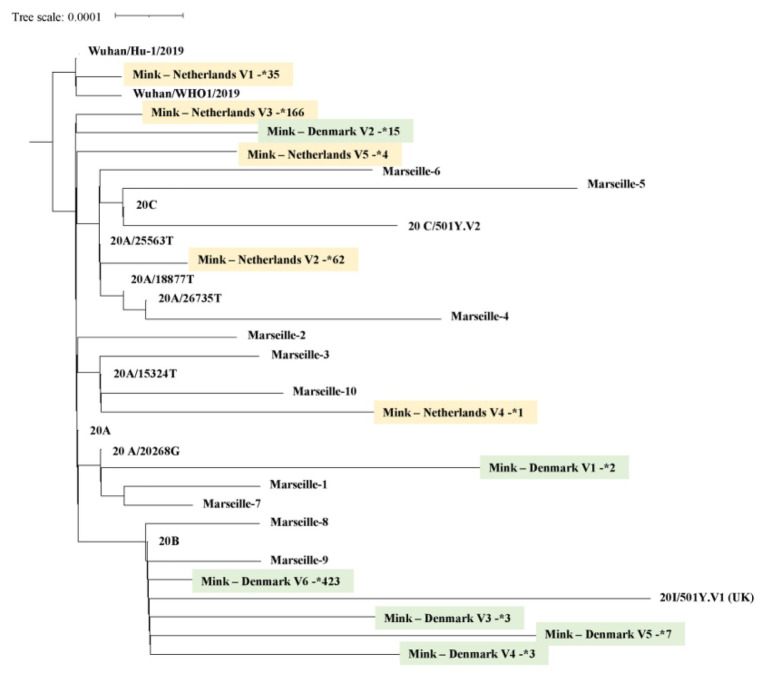
Figure 4.
Mink SARS-CoV-2 virus phylogeny. A total of 744 SARS-CoV-2, selected from GISAID (https://www.gisaid.org/), were integrated in a phylogenetic analysis. All genomes were aligned by using MAFFT version 7 (Katoh and Standley, 2013). A phylogenetic tree was reconstructed by using IQ-TREE with the GTR model with ultra-fast bootstrap of 1,000 repetitions (Minh et al., 2020). Sequences of mink from the Netherlands are highlighted in yellow, those from Denmark in green. The number next to the star is the number of genomes available for each mink SARS-CoV-2 genotype.