Figure 2. PIF1 is required for BIR to repair DSBs at broken forks to promote replication restart.
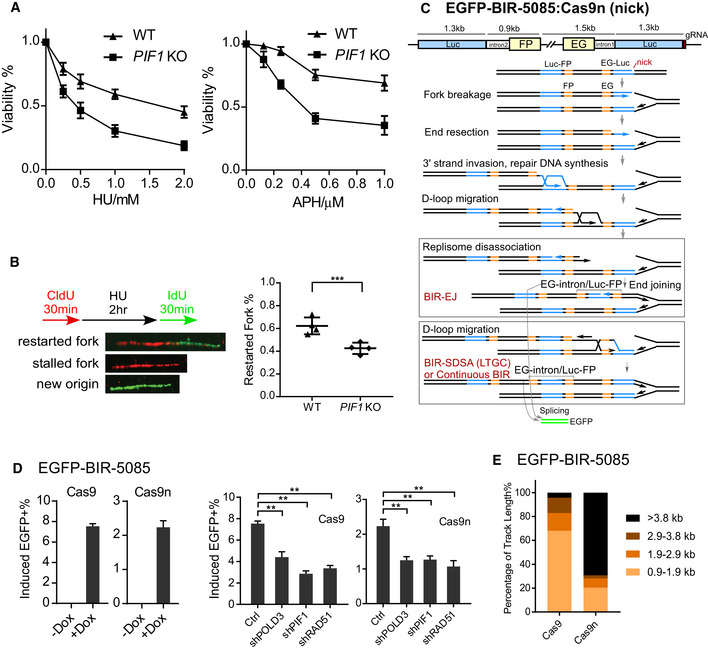
- U2OS WT or PIF1‐KO cells were treated with the indicated concentrations of HU (left) or APH (right) for 72 h, and the cell viability assay was performed.
- U2OS WT or PIF1‐KO cells were labeled with CldU for 30 min followed by incubation with 2 mM HU for 2 h and then IdU for another 30 min. Labeled cells were processed for DNA fiber analysis. Representative images of stalled or restarted forks and forks with new origin firing were shown (left). The percentage of restarted forks was quantified by analyzing of 110–130 fibers for each experiment (right). Experiments were repeated four times for each sample.
- Schematic drawing of the EGFP‐BIR‐5085 reporter and the repair steps leading to the repair product expressing EGFP after Cas9n/sgRNA‐5085 expression.
- U2OS (EGFP‐BIR‐5085) cell lines carrying Dox‐inducible Cas9/sgRNA‐5085 (Dox‐Cas9) or Cas9n/sgRNA‐5085 (Dox‐Cas9n) were incubated with or without Dox (5 µg/ml) and assayed by FACS analysis 2 days later (left). U2OS (EGFP‐BIR‐5085, Dox‐Cas9 or Dox‐Cas9n) cells expressing shRNAs RAD51, POLD3, and PIF1 or shRNA vector (Ctrl) were incubated with 5 µg/ml Dox, and FACS analysis was performed after 2 days (right).
- Track length of single EGFP‐positive clones derived from U2OS (EGFP‐BIR‐5085) cells after Cas9/sgRNA‐5085 (n = 47) or Cas9n/sgRNA‐5085 (n = 39) expression was analyzed by sequencing of the PCR products from genomic DNA covering the repair junctions.
Data information: Error bars represent the standard deviation (SD) of at least three independent experiments. Significance of the differences was assayed by two‐tailed non‐paired parameters were applied in Student's t‐test. The P value is indicated as **P < 0.01, ***P < 0.001.
