-
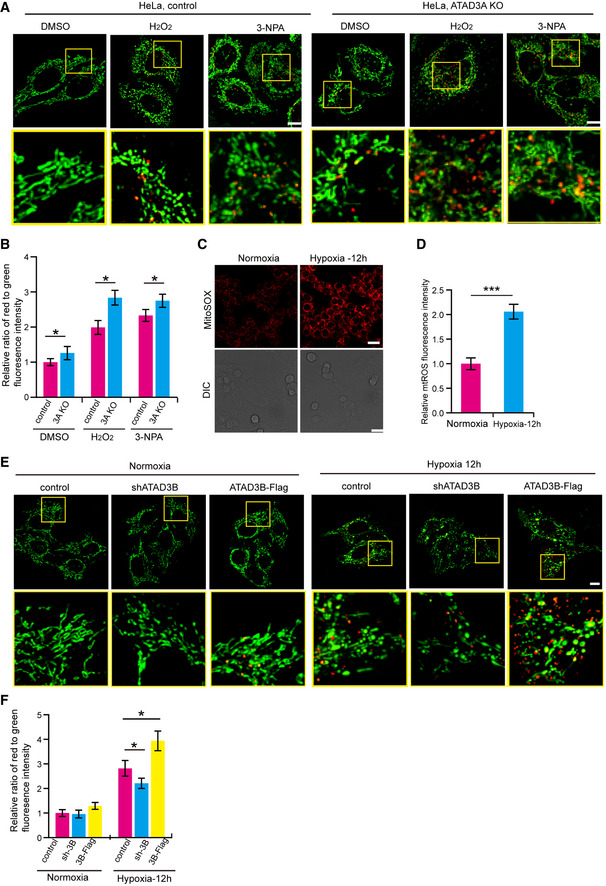
A
Control or ATAD3A KO HeLa cells stably expressing mito‐Keima were treated with DMSO, 4 mM 3‐NPA, or 200 µM H2O2 for 2 h. Then, cells were imaged with 458 nm (measuring mitochondria with a neutral pH) and 561 nm (measuring mitochondria with an acidic pH) laser excitation for mito‐Keima by confocal microscopy. Scale bar, 10 µm.
-
B
Quantification of the relative ratio of red to green fluorescence intensity (561 nm/458 nm) of the cells described in (A). Data are presented as mean ± SD (n = 3 independent experiments, 20 cells per experiment), and statistical significance was assessed by two‐tailed Student’s t‐test, *P < 0.05.
-
C, D
HeLa cells were cultured in normoxia or hypoxia (1% O2) for 12 h. Then, cells were stained with mitoSOX and analyzed by confocal microscopy (C). DIC, Differential Interference Contrast. Quantification of the MitoSOX fluorescence intensity was shown in the right panel (D). Data are presented as mean ± SD (n = 3 independent experiments, 20 cells per experiment), and statistical significance was assessed by Student’s t‐test, ***P < 0.001.
-
E
HeLa cells stably expressing mito‐Keima were infected with shATAD3B or ATAD3B‐Flag. Then, cells were cultured in normoxia or in hypoxia (1% O2) for 12 h. Cells were imaged with 458 nm (measuring mitochondria with a neutral pH) and 561 nm (measuring mitochondria with an acidic pH) laser excitation for mito‐Keima by confocal microscopy. Scale bar, 10 µm.
-
F
Quantification of the relative ratio of red to green fluorescence intensity (561 nm/458 nm) of the cells described in (A). Data are presented as mean ± SD (n = 3 independent experiments, 20 cells per experiment), and statistical significance was assessed by two‐tailed Student’s t‐test, *P < 0.05.