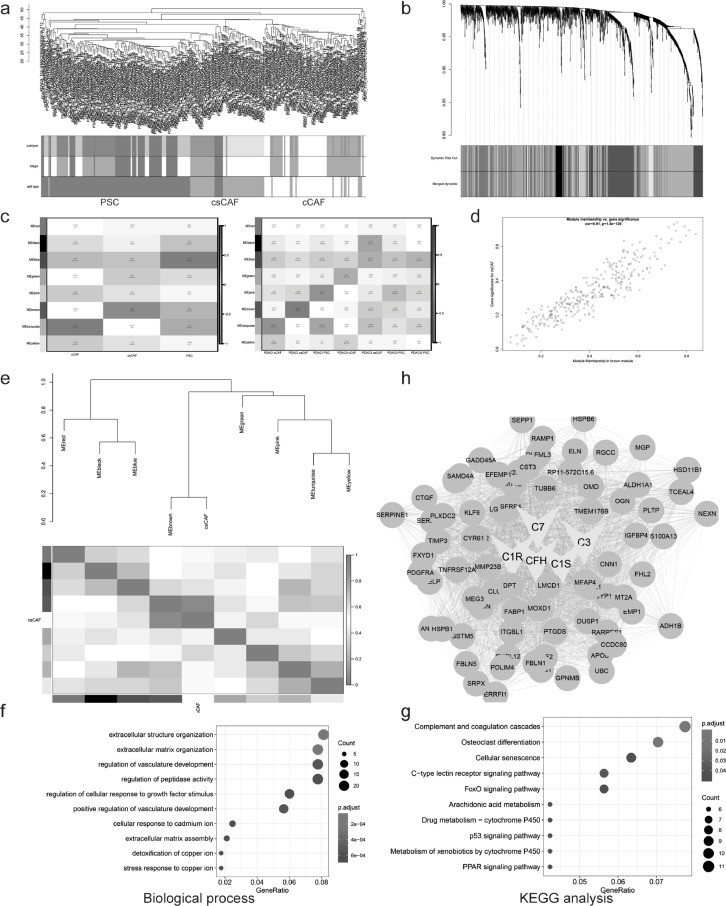
Fig. 6.
Gene-network module of csCAF were identified by WGCNA. (a) Clustering dendrogram of samples based on Euclidean distance. (b) Clustering dendrogram of genes, with dissimilarity based on topological overlap using Dynamic Tree Cut algorithm, together with assigned merged module colors and the original module colors. (c) The heatmap of module-trait associations. Each row corresponds to a module eigengene, column to CAF subsets, based on cell type in the left panel, subtype in the right panel. In addition, each box contained the corresponding correlation and p-value. (d) A scatter plot of Gene Significance (GS) for csCAF vs Module Membership (MM) in the brown module. There is a highly significant correlation between csCAFs and brown module. (e) Visualization of eigengene network representing the relationships among modules and csCAFs. The upper panel showed a hierarchical clustering dendrogram of the eigengenes, the lower panel showed the eigengene adjacency. (f-g) Gene ontology and KEGG analysis showing biological process (f) and pathway (g) terms for genes in brown module (csCAFs hub genes). (h) Visualization of network connections among the most connected genes in the brown module representing csCAFs using Cytoscape software.