FIGURE 3.
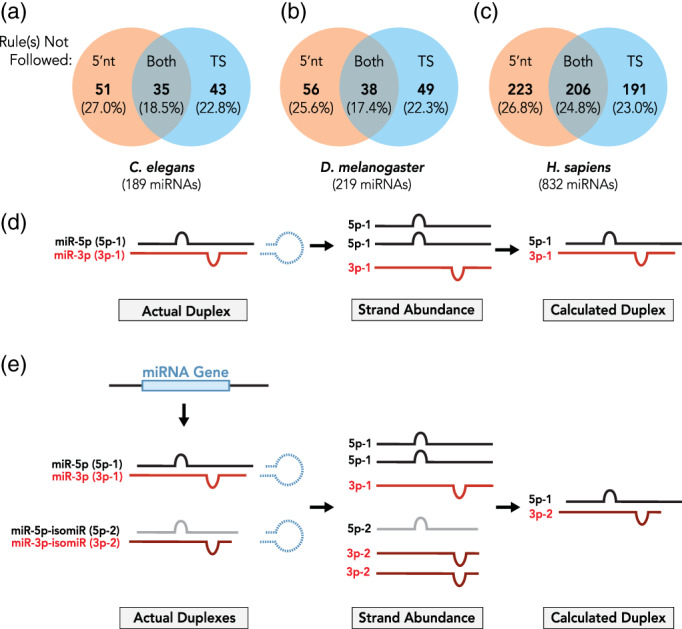
Not all miRNAs follow the rules. Shown are the percentages of miRNAs that do not follow the 5′ nucleotide selection rule, thermodynamic duplex‐end stability rule, or either rule in (a) Caenorhabditis elegans (n = 189 miRNAs), (b) Drosophila melanogaster (n = 219), and (c) humans (n = 832). miRNA sequences were obtained from miRBase (Release 22.1) and analyzed for duplex formation using RNAduplex (ViennaRNA package v2.4.14) with default parameters. The RNA secondary structure represented in dot‐matrix format was first examined for unpaired, overhanging bases at each duplex end. To determine duplex‐end stabilities, four terminal base pairs at each duplex end were loaded into RNAduplex for terminal minimum free energy (MFE) calculation. ΔΔG for duplex ends was calculated as follows: ΔΔG = Δguide − Δpassenger, where Δguide was the four base‐pair MFE value for the 5′ end of the guide miRNA and Δpassenger was the four base‐pair MFE for the 5′ end of the miRNA*. miRNAs were not considered to follow the 5′ nucleotide selection rule if the 5′ nucleotide of the passenger strand was identical to or more favorable than the 5′ nucleotide of the guide strand. The preferred 5′ nucleotide was defined as U > A > C > G (Frank et al., 2010). miRNAs with equal or higher thermodynamic stability on the guide end of the duplex compared to the passenger end of the duplex were not considered to follow the thermodynamic stability rule. (d) miRNA reads observed in small RNAseq experiments (“Strand abundance”) are thought to originate from the same duplex. (e) Hypothetically, for any given miRNA, miRNA reads observed in small RNAseq experiments may originate from distinct duplexes generated through alternative processing
