Figure 1. SFXN1, an integral inner mitochondrial membrane protein, is a TIM22 complex substrate.
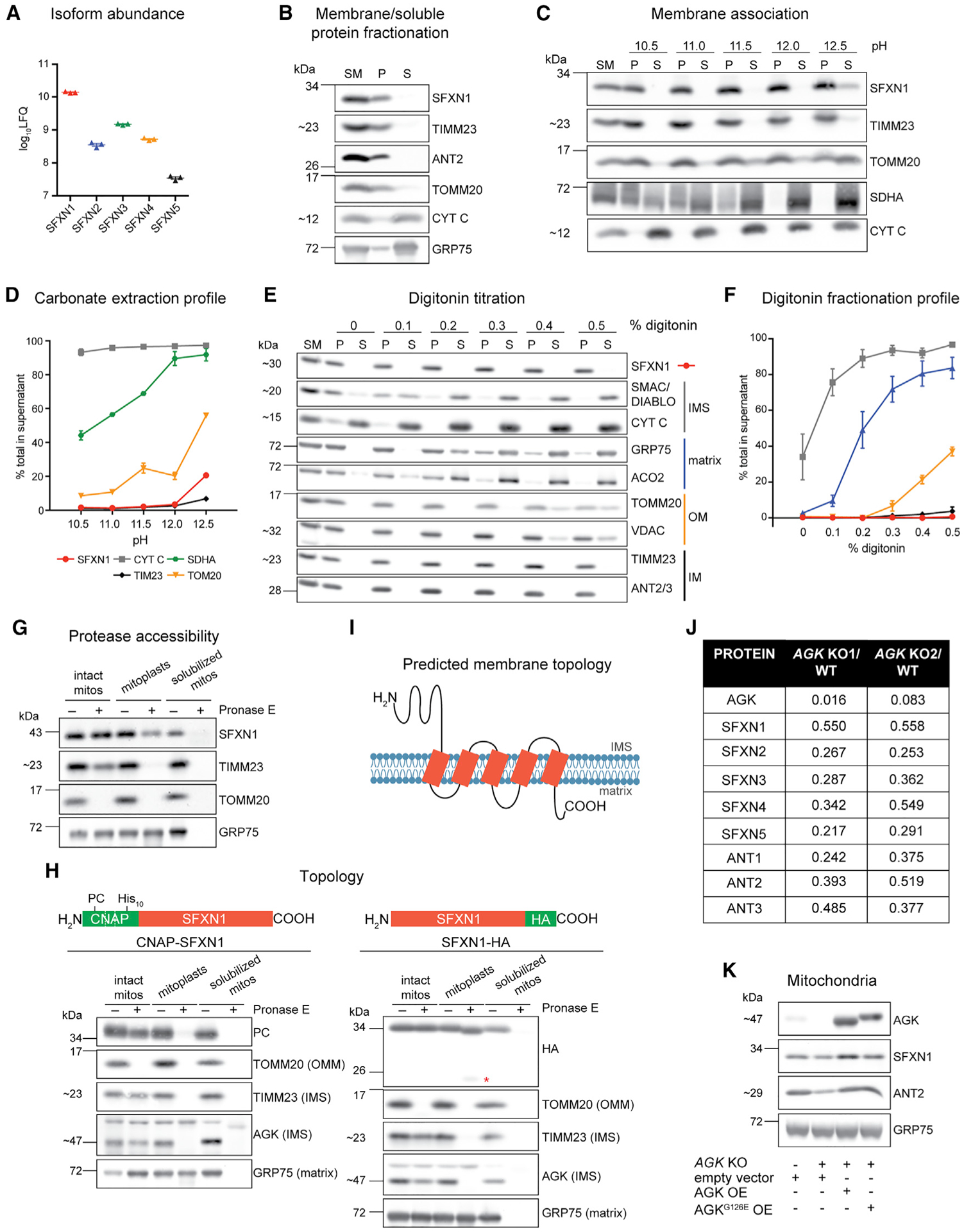
(A) Relative protein abundance of SFXN isoforms in HEK293 mitochondria as, determined by mass spectrometry and label-free quantification (LFQ) (mean ± SEM, n = 3).
(B) Sonication and centrifugation of mitochondria to separate membrane-bound from soluble proteins. SM, starting material; P, pellet; S, supernatant.
(C) Carbonate extraction of mitochondrial membrane proteins to distinguish between peripheral (appear in S) and integral (remain mostly in P) proteins.
(D) Band intensities of P and S fractions in (C) were quantified and plotted as % of protein released in the supernatant (mean ± SEM, n = 3).
(E) Digitonin titration for fractionation of mitochondrial subcompartments. Equal volumes of P and S fractions were analyzed.
(F) Band intensities of P and S fractions (E) were quantified. Average band intensity of representative mitochondrial proteins from each subcompartment was plotted as % of protein released in the supernatant (mean ± SEM, n = 3).
(G) Submitochondrial localization of endogenous SFXN1. HEK293 mitochondria were osmotically ruptured to yield mitoplasts or solubilized with sodium deoxycholate. Samples were treated with Pronase E where indicated.
(H) Submitochondrial localization of tagged SFXN1. HEK293 mitochondria lacking endogenous SFXN1 and expressing CNAP-SFXN1 or SFXN1-HA were processed as in (G). *, matrix-protected fragment.
(I) Predicted membrane topology of SFXN1 based on (H).
(J) Proteomic analysis of AGK KOs versus WT. Shown are relative protein amounts of SFXN and ANT isoforms in the presence or absence of AGK.
(K) Mitochondria from AGK KOs rescued with AGK, AGKG126E, or empty vector were resolved by SDS-PAGE and immunoblotted for the indicated proteins.
