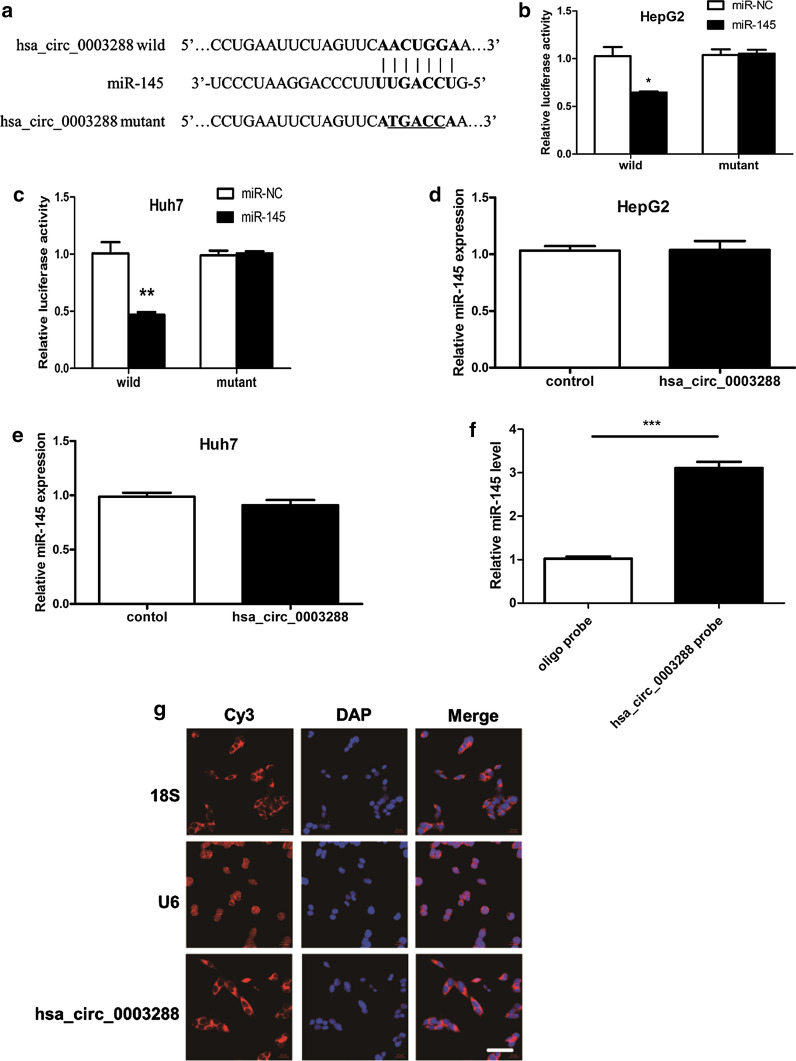
Fig. 2.
Hsa_circ_0003288 bind directly to miR-145 in HCC cells. a Predicted target sequences in hsa_circ_0003288 binding to miR-145. b, c Wild type or mutated hsa_circ_0003288 transfected into HepG2 or Huh-7 cells with miR-145 or negative control (miR-NC). Relative luciferase activity was determined 48 h post-transfection. U6 was used as the internal control. d, e Endogenous miR-145 levels in HepG2 and Huh-7 cells after transient overexpression of hsa_circ_0003288. f Cell lysates from HepG2 cells were incubated with biotinylated probes against hsa_circ_0003288; miR-145 enrichments was determined by qRT-PCR after pull-down. Results revealed that miR-145 was abundantly pulled down by the hsa_circ_0003288 probe in HepG2 cells. g The subcellular distribution of hsa_circ_0003288 was visualized by RNA Fluorescent in situ hybridization (FISH) in HepG2 cells. 18S was the positive control for cytoplasm, and U6 was the positive control for the nucleus. Scale bar: 20 μm. *P < 0.05; **P < 0.01