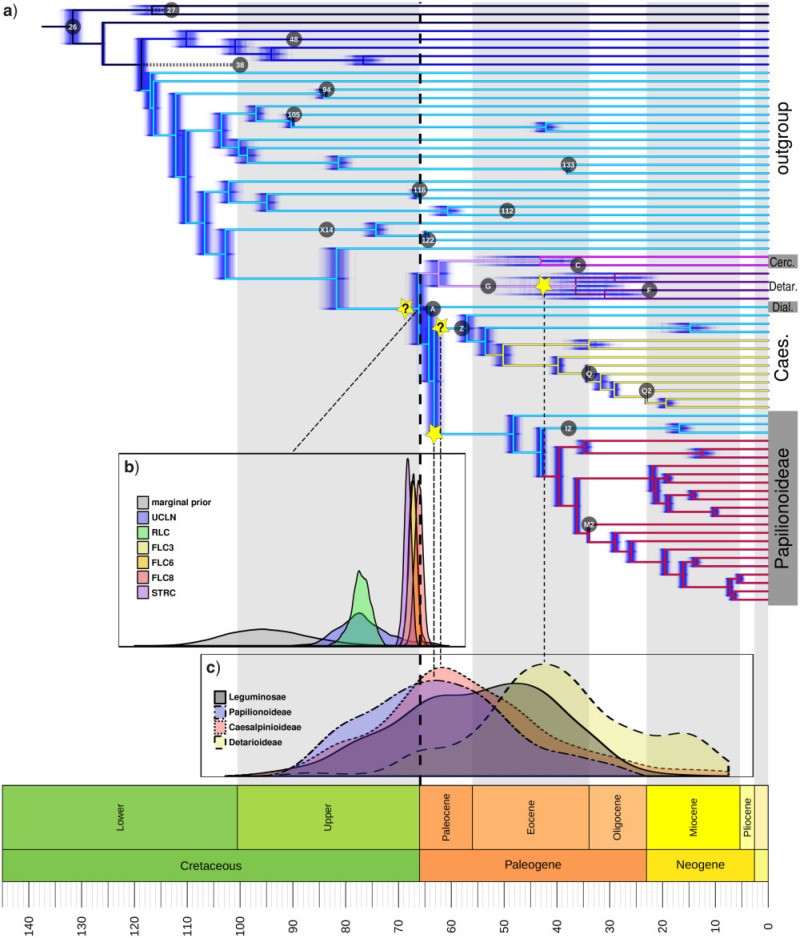
Figure 5.
The origin of the legumes is closely associated with the KPB. a) Chronogram estimated with eight fixed local clocks (FLC8 model) in BEAST, with the clock partitions indicated by colored branches, from an alignment of 36 genes selected as both clock-like and highly informative and hence well-suited for dating analyses. Blue shading represents 500 post-burn-in trees (“densitree” plot) indicating posterior distributions of node ages. Yellow stars indicate putative legume WGD events, the placement of a putative allopolyploid event is equivocal and is indicated by two stars labeled with question marks (one on the stem lineage of the family and one on the stem lineage of Caesalpinioideae because the time-scaling analysis of gene duplications presented in (c) is based on this subfamily). Labeled circles indicate placements and ages of fossil calibrations listed in Table 1. Note that fossil A is placed on the legume stem node but postdates the median crown age estimates for the family and is therefore not plotted on the legume stem lineage (similar for fossils 27 and 38). b) Prior and posterior distributions for the crown age of legumes under different clock models, as indicated in the legend. c) Density plots of age estimates for duplication nodes in gene trees, for all duplications that mapped onto the legume crown node in the Notung analysis in gray and for duplications in the three well-sampled subfamilies Papilionoideae, Caesalpinioideae, and Detarioideae as indicated in the legend.