FIG. 5.
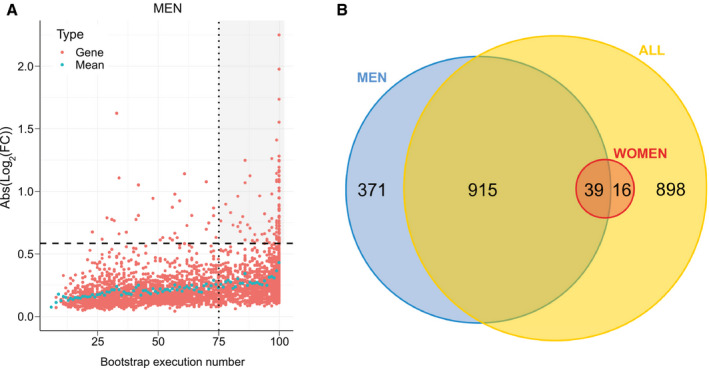
Identification of reliable DE genes. (A) The absolute log2FC of DEGs was computed for the men learning cohort ( , n = 85). Each significantly DEG (FDR < 10%) is represented by a red dot. Gene reliability is established by the number of bootstrap runs for which the gene remains significantly DE (75%). Blue dots represent the mean absolute log2FC for a given bootstrap run count. Dashed line, FC = 1.5; dotted line, occurrence = 75. The gray‐shaded area includes reliable DEGs (FC > 1.5) with occurrences ≥75. (B) Number of reliably identified DEGs between NoNASH and NASH groups (men [blue], women [red], and all patients [yellow]).
