Figure 4.
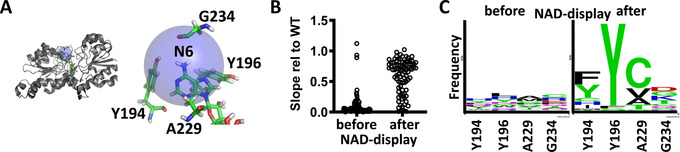
CaBoFDH SpliMLiB library design and screening by NAD‐display. A) On the left, the structure of CaBoFDH (PDB 5DN9) is shown with the targeted sites, Y194, Y196, A229 and G234, depicted in green and the 5 Å radius around the exocyclic nitrogen of adenine shown as a blue transparent sphere. On the right, the targeted residues and adenine are depicted in greater detail. Each of these residues was varied with codons encoding 12 different amino acids (Table S5), resulting in SpliMLiB library size of 20 736. B) Secondary screening of bacterial lysate before (input) and after (sorted output) flow cytometric sorting of beads. Progress curve slopes were normalised to wild‐type CaBoFDH lysate activity measured with 10 mM NAD+ and 10 mM sodium formate in a buffer consisting of 20 mM Tris‐HCl (pH 8) and 100 mM NaCl. C) Sequence logo depicting the frequency of amino acids encountered at each position for a total of 36 clones sequenced each for the beads before and after sorting.
