Figure 2.
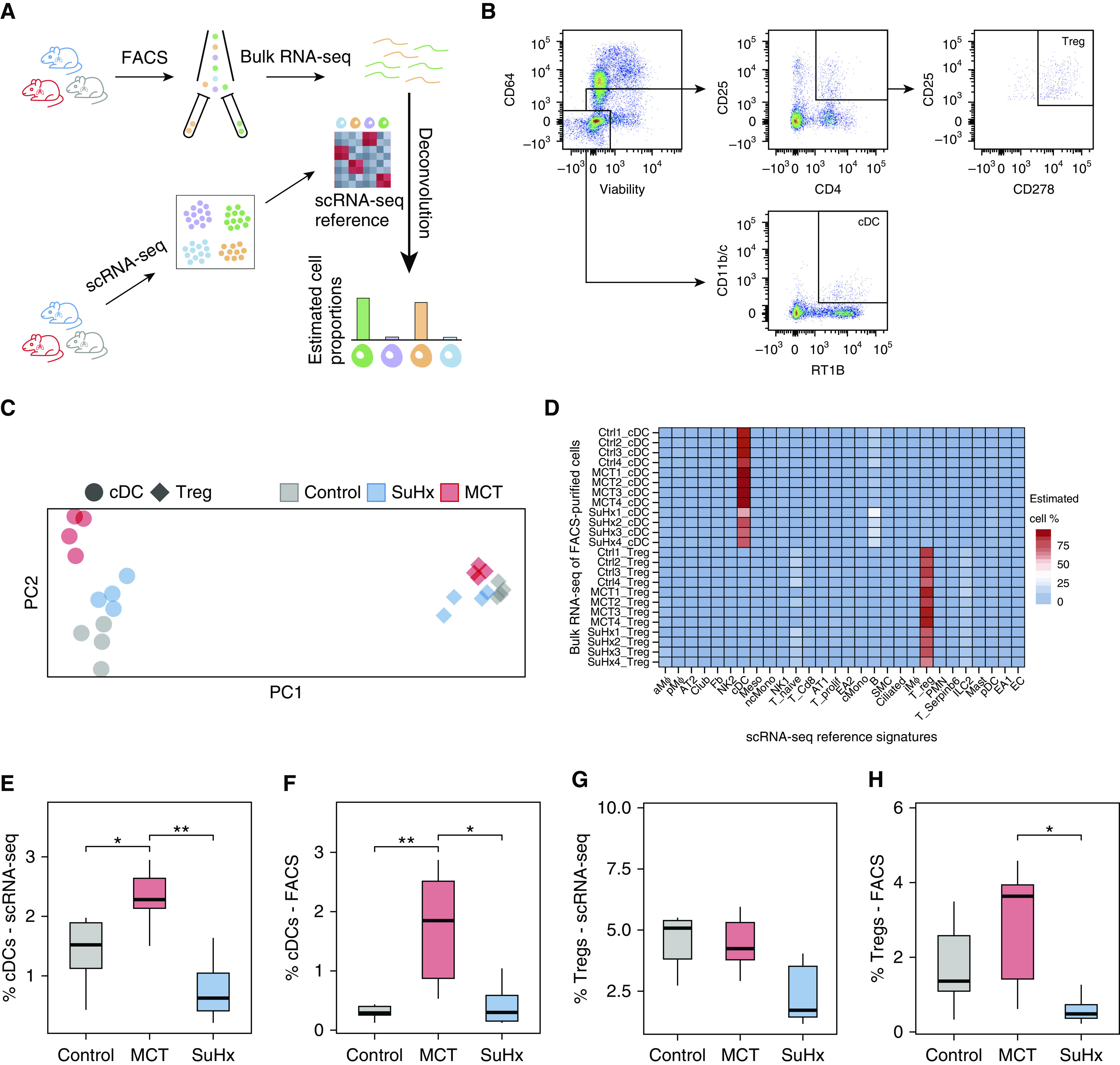
Fluorescence-activated cell sorting (FACS) and bulk RNA sequencing (RNA-seq) validate single-cell RNA-seq (scRNA-seq) cell-type identities and proportions. (A) Schematic showing the study design for deconvolution. The lungs from a separate set of Sugen-hypoxia (SuHx) (blue), monocrotaline (MCT) (red), and control (gray) rats (n = 4/group) underwent FACS using canonical cell-surface markers for two specific cell types, conventional dendritic cells (cDCs) and regulatory T cells (Tregs), after which bulk RNA-seq was performed on sorted cells to validate the cell-type identities in our scRNA-seq data using cell-type deconvolution of FACS-purified transcriptomes with CIBERSORTx (Stanford University). (B) Gating strategy for the isolation of CD64−, CD11b/c+, and RT1B+ cDCs and CD4+, CD25+, and CD278+ Tregs after gating for singlets and live cells. (C) Principal component (PC) analysis plot showing the clustering of bulk RNA-seq transcriptomes based on cell type (cDCs as circles and Tregs as diamonds) and disease condition. The first and second PCs (PC1 and PC2) explained 82% and 3% of the variance, respectively. (D) Heatmap showing deconvolution results using CIBERSORTx, in which relative proportions of cell types were estimated in each bulk RNA-seq sample of flow-sorted cells using a gene-expression signature for each cell type derived from scRNA-seq. High specificity for the correct cell types as identified by scRNA-seq was noted (83 ± 11% for cDCs; 77 ± 6% for Tregs). (E–H) FACS-determined relative cell-type proportions between disease models and the control model (F and H) showed a similar pattern to that of our scRNA-seq results (E and G). A significant increase in cDCs was noted in MCT compared with control rats. Furthermore, both methods consistently showed no significant changes in the number of cDCs in the SuHx model compared with the control model. The number of Tregs was also unchanged using either method in both disease models when compared with the control model. Wilcoxon rank-sum test: *P < 0.05 and **P < 0.01. FACS = fluorescence-activated cell sorter.
