Figure 5.
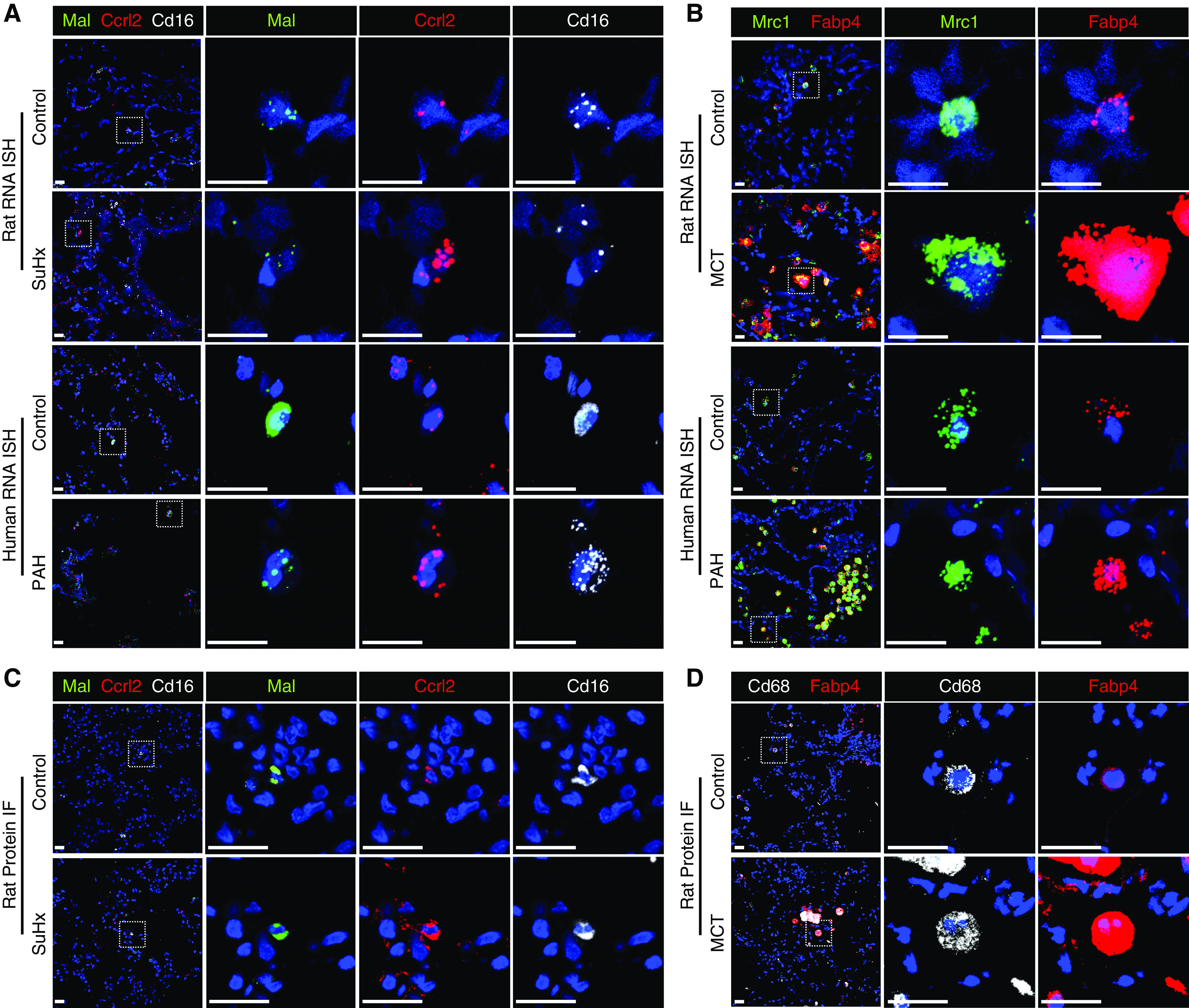
RNA ISH and immunofluorescence (IF) validate select differentially expressed genes. (A and B) The upregulation of Ccrl2 (red) in Sugen-hypoxia (SuHx) nonclassical monocytes (ncMonos) from single-cell RNA sequencing (scRNA-seq) was observed by RNAscope (Advanced Cell Diagnostics) in SuHx rats and patients with PAH (A) and by IF in SuHx rats (B). ncMonos were defined as cells positive for both Cd16 (white) and Mal (green). We chose Mal for double-labeling because it was the top marker gene specific for ncMonos in our data. (C and D) The upregulation of Fabp4 (red) in MCT alveolar macrophages (aMΦs) from scRNA-seq was demonstrated, in which aMΦs were defined as cells positive for Mrc1 (green) for rat and human RNAscope (C) or Cd68 (white) for rat IF (D). Both Mrc1 and Cd68 are canonical markers and were cell type–specific markers for aMΦs. The cell nuclei are labeled with DAPI (blue). Scale bars, 20 μm. ISH = in situ hybridization; MCT = monocrotaline; PAH = pulmonary arterial hypertension.
