Figure 1.
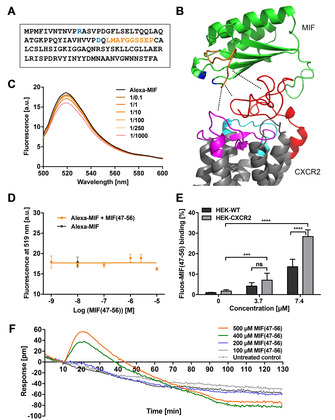
Interaction of peptide MIF(47–56) with CXCR2. A) Sequence of human MIF. Residues of the pseudo‐(E)LR motif (blue) and MIF sequence 47–56 (orange) are highlighted. B) Structure model of the complex between human MIF (green; PDB ID: 3DJH:A) and CXCR2 (gray; structure predicted by Phyre2 [11] and as predicted by protein–protein docking in PatchDock/FireDock for visualization purposes [12] ). The MIF sequence 47–56 is highlighted in orange, the pseudo‐(E)LR motif is depicted in blue; for CXCR2, the N‐domain (red), and parts of ECL1 (cyan) and 2 (magenta), that is, the regions that have been suggested to contribute to the interface with MIF, are also color‐coded. Dotted lines indicate interaction contact points. C), D) Fluorescence spectroscopic titrations of Alexa‐488‐labeled rMIF (Alexa‐MIF, 10 nM) with increasing concentrations of MIF(47–56) (0.1‐ to 1000‐fold molar excess). C) Fluorescence spectra of the various mixtures and of Alexa‐MIF alone recorded between 500 and 600 nm are shown. D) The fluorescence emission at 519 nm was plotted against the peptide concentration (three titration experiments, mean±SD). E) Binding of Fluos‐MIF(47–56) (3.7 or 7.4 μM) to CXCR2, stably expressed on HEK293 cells in comparison to non‐transfected wild‐type HEK293 cells. The mean fluorescence intensity (MFI) was measured by flow cytometry and intensities normalized to the signal of non‐transfected control cells (n=4–7, mean±SD). Statistical significance is indicated: *** P<0.001, **** P<0.0001; ns, not significant; WT, wild type. F) Detection of MIF(47–56) binding to CXCR2 by label‐free dynamic mass redistribution (DMR) technology. HEK293‐CXCR2 transfectants were treated with MIF(47–56) (at 100, 200, 400, 500 μM, as indicated), and cellular responsiveness as a measure of binding was recorded at 30 s intervals for a total of 120 min.
