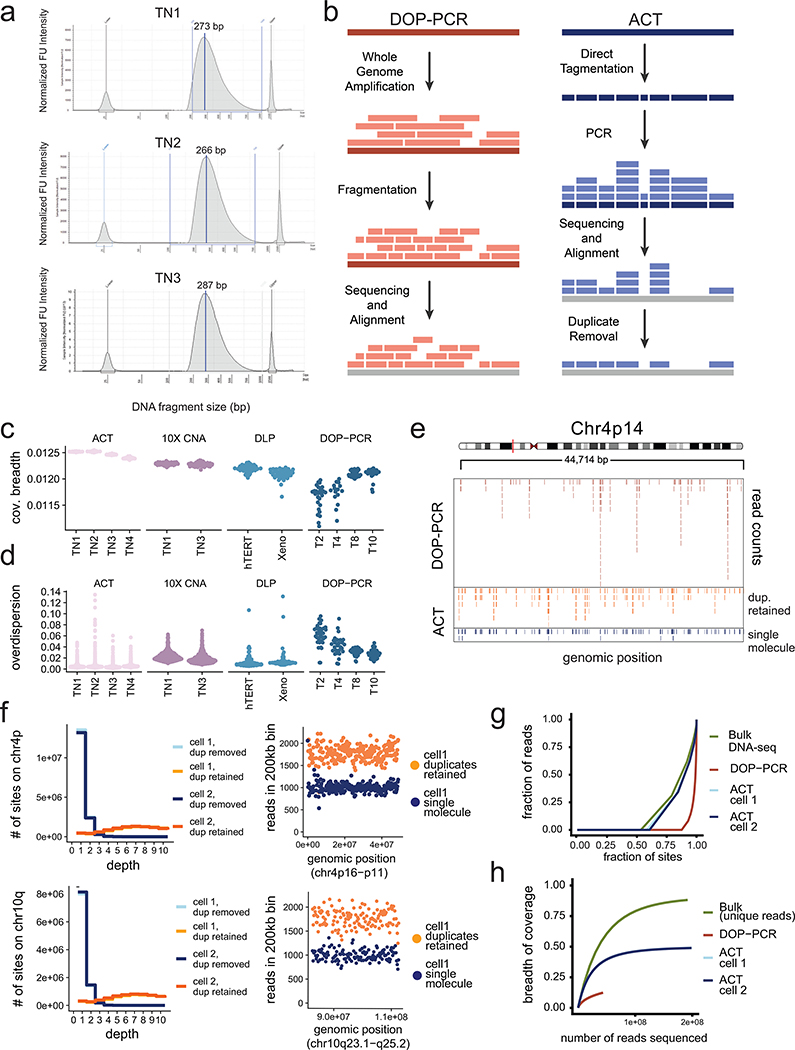
Extended Data Figure 1 – Technical Metrics and Performance of ACT.
(a) ACT single cell DNA library size distributions for TN1, TN2 and TN3 after pooling 384 cell libraries. (b) Schematic of using positional barcoding information to determine single-molecule information by tagmentation during ACT, compared to whole-genome amplification using DOP-PCR, where the original DNA fragmentation sites of single molecules cannot be resolved. (c) Breadth of coverage for sparse depth data from different scDNA-seq methods plotted by individual samples, using N=100 random cells per sample. (d) Overdispersion of bin counts for sparse depth data from different scDNA-seq methods plotted by individual samples, using N=100 random cells per sample. (e) Distribution of sequencing reads across a diploid region of chromosome 4p14 for a single SK-BR-3 cell sequenced by DOP-PCR compared to ACT, in which the PCR duplicates were retained or removed to obtain single-molecule data. (f) Distribution of sequencing reads across a diploid region of chromosome 4p (top panel) and 10q (bottom panel) for a single SK-BR-3 cell sequenced by DOP-PCR compared to ACT, with or without duplicate molecules retained. (g) Lorenz curves of coverage uniformity for ACT, DOP-PCR and one bulk DNA-seq data from SK-BR-3 single cells, downsampled to equal coverage depth. (h) Breadth of coverage as a function of pseudo-bulk reconstruction by combining multiple cells for ACT, DOP-PCR and bulk sequencing.