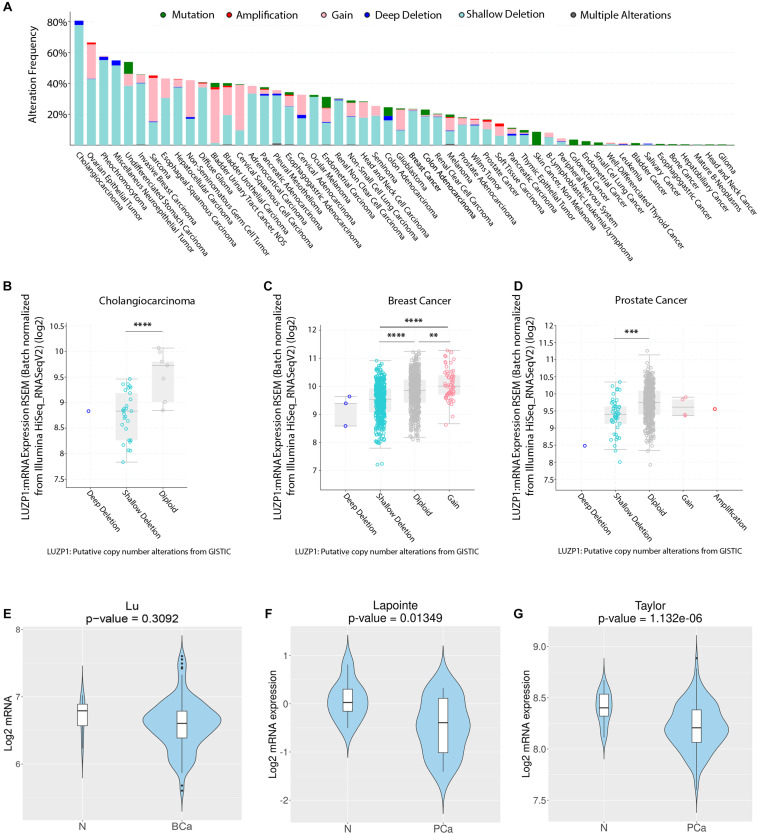
FIGURE 1.
Copy number alterations and aberrant LUZP1 gene expression in cancer. (A) Copy number alterations (CNA) (mutations-green; amplifications-red; gain-light pink; deep deletions-blue; shallow deletions-aquamarine; multiple alterations-gray) in different TCGA tumor types (n = 10,967). (B–D) LUZP1 mRNA expression (RNAseq) sorted by CNA in (B) Cholangiocarcinoma (n = 195), (C) Breast (n = 1,918), and (D) Prostate Cancer samples (n = 324). When n > 3, data were analyzed by One-way ANOVA or Kruskall-Wallis and the corresponding post hoc test. (E) Violin plots depicting the expression of LUZP1 between non-tumoral (N) and breast cancer specimens (BCa) in the Lu dataset (Lu et al., 2008). (F,G) Violin plots depicting the expression of LUZP1 between non-tumoral (N) and prostate cancer specimens (PCa) in Lapointe et al. (2004) (F) and Taylor et al. (2010) (G) datasets. The Y-axis represents the Log2-normalized gene expression (fluorescence intensity values for microarray data or sequencing read values obtained after gene quantification with RSEM and normalization using Upper Quartile in case of RNAseq). Student T-test was performed in order to compare the mean gene expression between two groups. ∗∗P < 0.01; ∗∗∗P < 0.001; ****P < 0.0001.