Fig 2. Comparison of the coverage of several genomic DNAs and NIPT single cells.
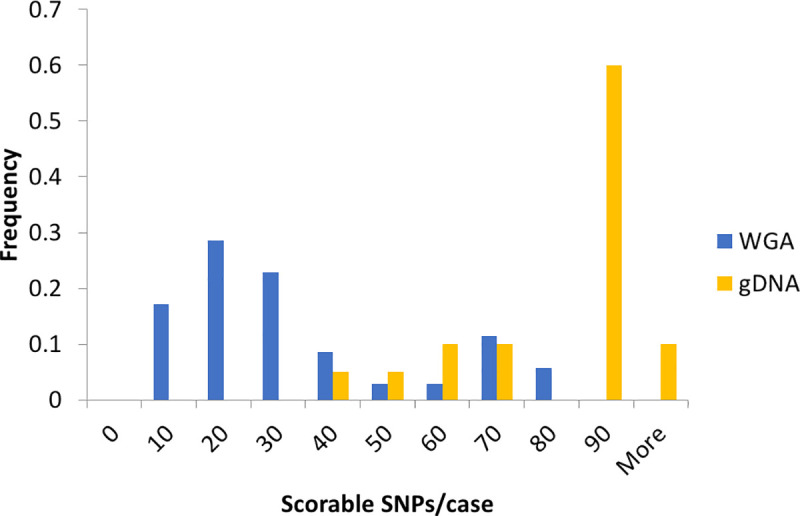
The count of samples with various scorable SNPs was normalized to the total number of samples of each group (gDNA n = 20, WGA n = 35). The scorable SNP cutoff was at least 5% minor allele frequency (MAF) and ten reads. Data include NIPT case numbers 946, 977, 982, 983, 984, 988, 989, 990, 991, 992, 993, 996, and 998 (clinical data in S8 File).
