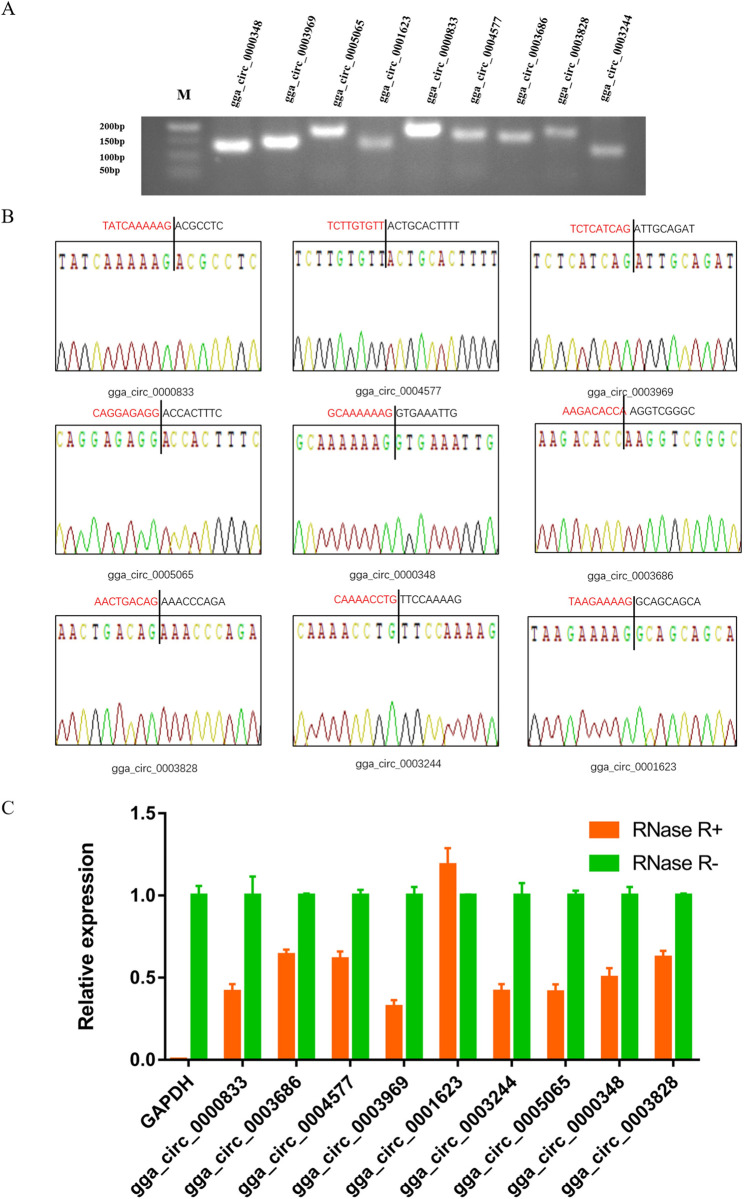
Fig 1. Verification of 9 randomly selected circRNAs.
(A) Electrophoretogram of the PCR products. M: marker 500. (B) The head-to-tail junctions of 9 circRNAs were confirmed by Sanger sequencing. The black line indicates the position of reverse splicing. The area marked with red (or black) is the end (or start) sequence of the circRNAs (junction). (C) qRT-PCR was used to detect the resistance of circRNA to RNase R digestion. GAPDH was used as a linearity control. Data are expressed as means ± SEM (n = 3).