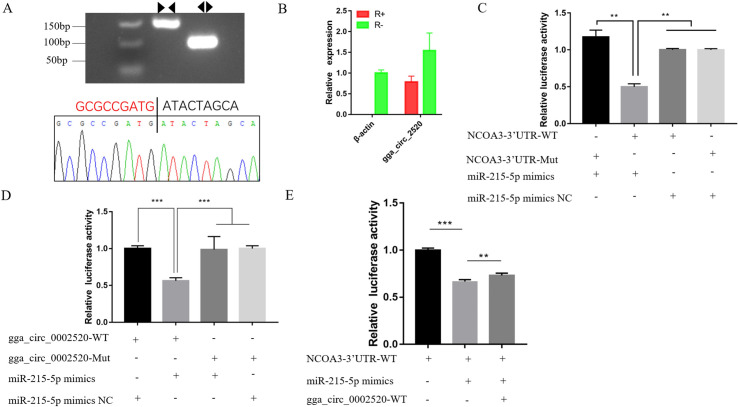
Fig 9. Experimental verification of the ceRNA gga_circ_0002520-miR-215-5p-NCOA3.
(A) Convergent and divergent primers amplify circRNAs in cDNA. The head-to-tail junctions of gga_circ_0002520 were confirmed by Sanger sequencing. The black line indicates the position of reverse splicing. (B) qRT-PCR was used to detect the resistance of gga_circ_0002520 to RNase R digestion. β-actin was used as a linearity control. Data are expressed as means ± SEM (n = 3). (C) Luminescence was measured after cotransfecting the wild-type or mutant sequence of NCOA3-3′UTR with miR-215-5p mimics (or mimics NC) in DF-1 cells. (D) Luminescence was measured after cotransfecting the wild-type or mutant linear sequence of gga_circ_0002520 with miR-215-5p mimics (or mimics NC) in DF-1 cells. (E) A double luciferase assay was performed in DF-1 cells 48 h after miR-215-5p mimic transfection with NCOA3-3′UTR-WT and gga_circ_0002520-WT.