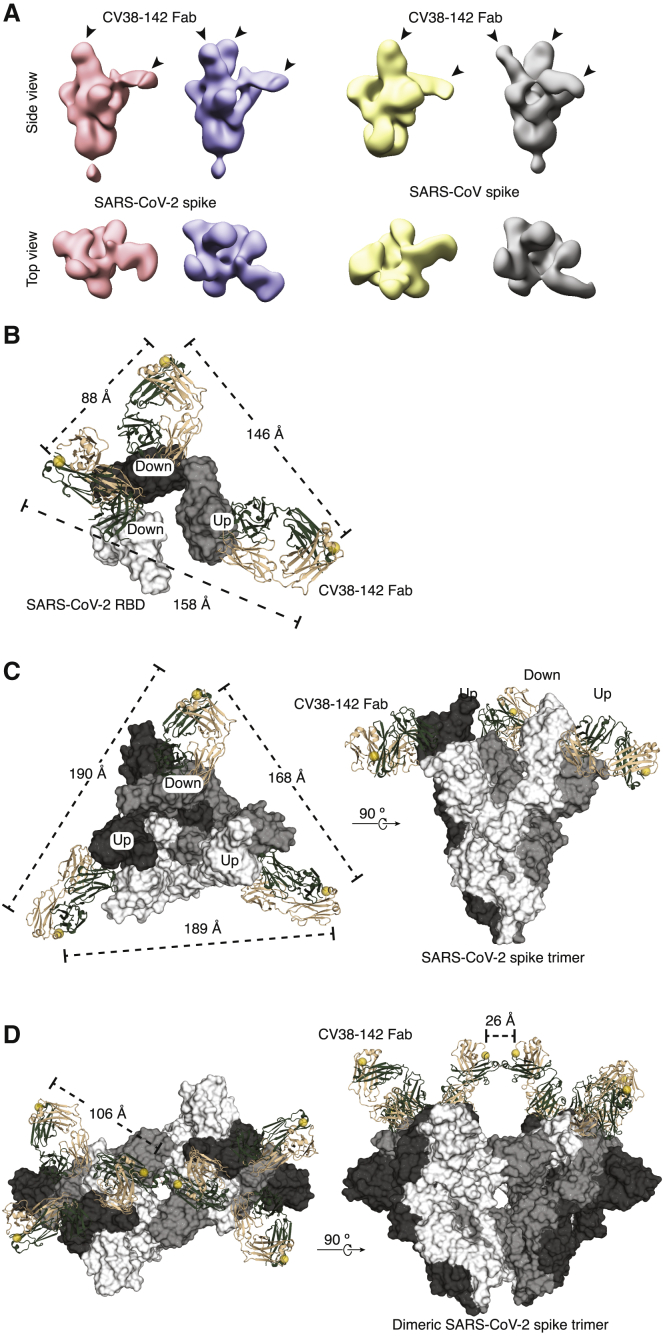
Figure 6.
CV38-142 Fab binding to SARS-CoV-2 and SARS-CoV spike trimers by nsEM
(A) CV38-142 Fab binding to spike trimers as observed by nsEM. Representative 3D nsEM reconstructions are shown of CV38-142 Fab complexed with the spike trimers with its RBDs in “up” and “down” states. The location of the bound CV38-142 Fabs are indicated by arrow heads. SARS-CoV-2 (pink) or SARS-CoV (yellow) spikes with at least one “up” RBD and one “down” RBD are bound by two CV38-142 Fabs. The spikes (pale blue to SARS-CoV-2 and gray to SARS-CoV) with RBD in the two “down,” one “up” states are bound by three Fabs. Other binding stoichiometries and conformations are show in Figure S6.
(B–D) C-terminal distances of CV38-142 Fab binding to spikes. The three RBDs (B) or three protomers (C and D) in the spike trimer are shown in white, gray, and dark gray, respectively. CV38-142 Fabs are shown in ribbon representation with heavy chain in forest green and light chain in wheat. The C termini of CV38-142 heavy chains are shown as spheres (yellow). Dashed lines represent distances among the various combinations of C-termini.
(B) nsEM fitting model. To measure the distances between C-termini of CV38-142 Fabs in nsEM data, the crystal structure of CV38-142 Fab + SARS-CoV-2 was fitted into the nsEM density in (A) (second from the left).
(C and D) Structural superimposition of CV38-142 Fabs onto the spike trimer, which is shown in surface representation. Alignment of CV38-142 Fab binding to the spike trimer with RBD in two “up,” one “down” state (PDB: 7CAI) (C) or to a dimeric spike trimer that is found in Novavax vaccine candidate NVAX-CoV2373 with RBD in “all-down” state (PDB: 7JJJ) (Bangaru et al., 2020) (D).
The (B–D) models represent various possibilities of CV38-142 binding to the spike protein on the viral surface.
See also Figure S6.