Abstract
MicroRNAs (miRNAs) are small noncoding RNAs (18–24 nt) and function in many biological processes in plants. Although Eucalyptus trees are widely planted across the world, our understanding of the miRNA regulation in the somatic embryogenesis (SE) of Eucalyptus is still poor. Here we reported, for the first time, the miRNA profiles of differentiated and dedifferentiated tissues of two Eucalyptus species and identified miRNAs involved in SE of Eucalyptus. Stem and tissue culture–induced callus were obtained from the subculture seedlings of E. camaldulensis and E. grandis x urophylla and were used as differentiated and dedifferentiated samples, respectively. Small RNA sequencing generated 304.2 million clean reads for the Eucalyptus samples (n = 3) and identified 888 miRNA precursors (197 known and 691 novel) for Eucalyptus. These miRNAs were mainly distributed in chromosomes Chr03, Chr05, and Chr08 and can produce 46 miRNA clusters. Then, we identified 327 and 343 differentially expressed miRNAs (DEmiRs) in the dedifferentiation process of E. camaldulensis and E. grandis x urophylla, respectively. DEmiRs shared by the two Eucalyptus species might be involved in the development of embryonic callus, such as MIR156, MIR159, MIR160, MIR164, MIR166, MIR169, MIR171, MIR399, and MIR482. Notably, we identified 81 upregulated and 67 downregulated miRNAs specific to E. camaldulensis, which might be associated with the high embryogenic potential. Target prediction and functional analysis showed that they might be involved in longevity regulating and plant hormone signal transduction pathways. Further, using the gene expression profiles, we observed the negative regulation of miRNA–target pairs, such as MIR160~ARF18, MIR396~GRF6, MIR166~ATHB15/HD-ZIP, and MIR156/MIR157~SPL1. Interestingly, transcription factors such as WRKY, MYB, GAMYB, TCP4, and PIL1 were found to be regulated by the DEmiRs. The genes encoding PIL1 and RPS21C, regulated by upregulated miRNAs (e.g., egd-N-miR63-5p, egd-N-miR63-5p, and MIR169,) were downregulated exclusively in the dedifferentiation of E. camaldulensis. This is the first time to study the miRNA regulation in the dedifferentiation process of Eucalyptus and it will provide a valuable resource for future studies. More importantly, it will improve our understanding of miRNA regulation during the somatic embryogenesis of Eucalyptus and benefit the Eucalyptus breeding program.
Keywords: Eucalyptus, miRNA, vegetative propagation, somatic embryogenesis, dedifferentiation, callus
Introduction
Eucalyptus, a highly diverse genus of the Myrtaceae family, is the most widely planted hardwood due to its increasing importance for fiber and energy in the world. Their remarkable adaptability coupled with fast growth and superior wood properties has driven their rapid adoption for plantation forestry in South America, Africa, Asia, Spain, Portugal, Middle Eastern countries, and North America (Durand-Cresswell et al. 1982). This genus consists of about 700 species, of which Eucalyptus globulus (E. globulus) and E. grandis are currently the most extensively planted species (Pappas et al. 2015). Natural regeneration of Eucalyptus mainly relies on seed and the breeding process is generally slow because of the length of the juvenile phase before flowering. Although natural hybridization of Eucalyptus has often been reported to gain some characteristics (e.g., cold resistance), artificial hybridization is still a means of producing a limited number of seeds and the selected hybrids must then be multiplied by vegetative propagation (air layering, grafting, stem cutting, and tissue culture) (Durand-Cresswell et al. 1982). During the in vitro tissue culture–induced organogenesis or somatic embryogenesis (SE), cells of explants are activated to dedifferentiate and re-enter the cell cycle and, in some cases, shoots or roots are also regenerated after callus formation (Ikeuchi et al. 2013; Pulianmackal et al. 2014). The first step of SE is the callus induction, which includes a number of characteristic events: dedifferentiation of cells, activation of cell division and reprogramming of cell physiology, metabolism, and gene expression patterns (Zimmerman 1993). The second step of SE is the plant regeneration, which is related to the ability to regenerate plant (also called as embryogenic potential) (Dinkova and Alejandri-Ramirez 2014). Some studies have been demonstrated to study the molecular mechanisms of SE in other plants, such as maize (Lopez-Ruiz et al. 2018), rice (Zhang et al. 2019), banana (Kumaravel et al. 2017), and Catalpa bungei (Liu et al. 2019). However, large is not clear about the molecular mechanisms in the SE of Eucalyptus.
MicroRNAs (miRNAs) are a class of small (∼21 nt) noncoding RNAs that function in regulating gene expression at posttranscriptional level (Ji et al. 2014; Pappas et al. 2015). They have been reported to function in multiple biological processes in plant, such as development, flowering, architecture, and response to biotic and abiotic stresses (Axtell et al. 2007). Some studies have reported miRNAs associated with SE and dedifferentiation in other plants. Sabana identified 4 upregulated (miR160a, miR166a, miR171a, and miR319b) and 6 downregulated (miR156f, miR167c, miR169a, miR319a, miR535a, and miR5179) miRNAs in the embryogenic compared with nonembryogenic calli of coconut (Sabana et al. 2020). miR166 was reported to be downregulated while its target gene ATHB15 was upregulated from the longan embryonic callus to the globular embryo stages (Zhang et al. 2020). Jin summarized the miRNAs targeting the phytohormone signaling pathways during SE, including miR156/miR157~SPL2,9,10,14, miR159~MYB33, miR390~TAS3, miR396~GRF, miR160~ARF10,16, miR165/166~HD-ZIPIII, miR167∼ARF6/8, and miR393~TIR1, AFB2 (Jin et al. 2020). Some studies have been demonstrated to identify miRNAs in Eucalyptus. For example, Levy found no mutual relationship between alternations in miR156 and miR172 expression in the adventitious root induction (Levy et al. 2014); Pappas reported the variable patterns of conservation and diversity with miRNAs across Myrtaceae species (Pappas et al. 2015); and Lin identified 386 novel miRNAs in E. grandis and predicted their target genes (Lin et al. 2018). These studies provide a basis of understanding the miRNA regulatory networks in Eucalyptus. However, little is known about the miRNA regulation in the SE of Eucalyptus.
Small RNA sequencing has been used to identify known and novel miRNAs in animals and plants. In Eucalyptus, it has been used to characterize the miRNAs and their targets (Pappas et al. 2015; Lin et al. 2018). Previously, we analyzed the transcriptome profiles of SE and identified genes involved in the SE of Eucalyptus (Xiao et al. 2020). In this study, we aimed to identify the miRNA profiles of differentiated (stem) and dedifferentiated (callus) tissues in E. camaldulensis (high embryogenic potential) and E. grandis x urophylla (low embryogenic potential). Also, we aimed to characterize differentially expressed miRNAs (DEmiRs) between differentiated and dedifferentiated tissues, which might be associated with the embryogenic potential of Eucalyptus. Our findings will provide a valuable resource for future studies and improve our understanding toward the molecular mechanisms during the SE and propagation development of Eucalyptus. The output of this study will benefit the breeding program of Eucalyptus in this field.
Materials and methods
Ethics approval
No specific permits were required for the described field studies. The location is not privately owned or protected in any way, and the field studies did not involve endangered or protected species.
Plants and tissue culture
The original seeds of E. camaldulensis (high regenerative ability, voucher ID: c0009) and E. grandis x urophylla (low regenerative ability, voucher ID: j0017) were obtained from the wild in 1984. The plants were confirmed by Prof. Dongyun Xiang (senior botanist) and maintained in the experimental fields of Guangxi Forestry Research Institute. The second generation of in vitro tissue culture–induced seedlings of E. camaldulensis and E. grandis x urophylla were maintained on the MS medium supplemented with 20 mg/l Ca(NO3)2, 0.5 mg/l 6-BA, and 0.1 mg/l IAA until 2–3 cm long for this project. The second to the third stems from the stem tip of the seedlings were obtained and cut into 0.3–0.5 cm segments. About 60 segments of each Eucalyptus species were then transferred onto the induction MS medium [supplemented with 20 mg/l Ca(NO3)2, 1 mg/l KT, and 0.5 mg/l 2,4-D] and maintained in dark at 28 ± 2°C for 10 days. Successful callus samples were used as dedifferentiated tissues. The induction experiment was replicated three times.
RNA isolation and quality control
Total RNA was isolated from the plant tissues (100 mg, in triplicates) using the TRIzol reagent (Invitrogen) according to the manufacturer’s protocol, as previously described (Ji et al. 2014). The quantity and quality of total RNA were evaluated by Agilent Bioanalyzer 2100 (Agilent Technologies).
Library preparation of small RNAs
Total RNA (1 µg) of each sample was used to construct the small RNA libraries. Initially, total RNA samples were fractionated on a 15% urea-PAGE gel electrophoresis and a band corresponding to small RNAs (18–30 bp) was excised. After small RNAs extracted by centrifugation, they were ligated with the adenylated 3′ adapter. Then, RT primer with barcode was used to anneal the 3′ adenylated adapter, followed by the ligation of 5′ adapter and the reverse transcript reaction. After the first strand cDNA synthesis, we amplified the product by 15 cycles and carried out another size selection (103–115 bp) from the gel. After gel purification, the PCR product was quantified by Qubit (Invitrogen) and a single strand DNA circle (ssDNA circle), which gave the final miRNA library, was made by pooling multiple samples together.
Small RNA deep sequencing
We first generated the DNA nanoballs (DNBs) with ssDNA circle by rolling circle replication to enlarge the fluorescent signals at the sequencing process, as previously described (Drmanac et al. 2010). Then, the DNBs were loaded into the patterned nanoarrays and single-end sequencing strategy (SE50) was performed on the BGISEQ-500 platform in BGI-Shenzhen.
Data analysis and miRNA identification
The raw reads were cleaned by SOAPnuke (v2.1.0) to remove low-quality reads and sequencing adaptors, as previously described (Liu et al. 2017). In order to identify miRNAs in the Eucalyptus genome, we first merged the clean reads of all samples into one file and filtered lowly expressed reads (<50 copies) (Li et al. 2017). The retained clean data were aligned to the Eucalyptus genome (http://plantgenie.org) using SOAP2 (Li et al. 2009) without mismatches and potential miRNA precursors were predicted using MIREAP (https://github.com/liqb/mireap), as described (Li et al. 2017). Then, potential miRNAs were searched against all the plant miRNA sequences in miRBase (v22.1). The family information of Eucalyptus miRNAs was manually identified using their similarities to known miRNAs in other species. miRNA clusters were identified using a 10,000 bp window screening in the genome. miRNAs not aligned to other known miRNAs were considered as novel miRNAs.
Expression profile of small RNAs
Before miRNA expression was profiled, we annotated other kinds of small RNAs in each sample. First, ribosomal RNA (rRNA), transfer RNA (tRNA), small cytoplasmic RNA (scRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and signal recognition particle RNA (srpRNA) were annotated by mapping the clean reads to NCBI GenBank and Rfam (v14.0) databases (Kalvari et al. 2021). Then, mRNA fragments were identified by mapping the clean reads to the Eucalyptus mRNA sequences. After the reads aligned to the rRNA, tRNA, scRNA, snRNA, snoRNA, and srpRNA were removed, we aligned the retained clean reads of each sample to all the miRNA precursors identified before using BLAST (no mismatch). Reads aligned to each mature miRNA were zero counted. Normalization of miRNA expression was performed using the transcripts per million reads (TPM) method and total clean reads were used as background.
Differential expression analysis
We performed differential expression analysis of miRNAs using edgeR, as previously described (Chen et al. 2019). Raw counts of all miRNAs were used as input for edgeR. Some cutoffs were applied to the expression levels and statistical values to select differentially expressed miRNAs, such as TPM > 5, log2 fold change (log2fc) > 1 or log2fc < −1, P-value < 0.05 and false discovery rate <0.1.
miRNA target prediction
Target prediction of the Eucalyptus miRNAs was performed using three software, including psRobot (Wu et al. 2012), TarHunterL (https://github.com/XMaBio/TarHunterL), and TargetFinder (https://github.com/carringtonlab/TargetFinder). Considering the miRNA–mRNA interaction sites are conserved, a target gene identified by all three software were considered to be the target of an miRNA. The gene sequences of Eucalyptus were obtained from The Plant Genome Integrative Explorer (http://plantgenie.org) and their annotation were obtained from the NCBI platform.
Functional analysis
We first annotated the Eucalyptus genes by mapping them to the KEGG and GO databases. Then, the enrichment of GO terms and pathways by the miRNA target genes were identified using two statistical values—P-value (calculated by Fisher’s exact test, <0.05) and q-value (calculated by the R package “qvalue,” <0.05), as previously described (Wei et al. 2017).
Quantitative real-time PCR
We applied the poly-A tail extension method to the quantitative real-time PCR (qRT-PCR) experiment to confirm the expression levels of miRNAs identified in this study. A total of seven miRNAs (miR156a-3p, miR482b-3p, miR390a-3p, miR396d-3p, N-miR1-5p, N-miR6-3p, and N-miR18-5p) were randomly selected and the actin gene was used as internal control. Primer sequences for the miRNA candidates and internal control were predicted and synthesized at BGI-Shenzhen (Shenzhen, China). The qPCR procedures were the same as a previous study with some modifications (Shi and Chiang 2005). Briefly, we used the miRNA First Strand cDNA Synthesis (Tailing Reaction) reagent (B532451, Sangon Biotech, Shanghai, China) to add the poly-A tail to the RNA and reverse transcription. Then, the cDNA product (diluted 10 times) was used for the qRT-PCR experiment with the miRNAs qPCR Kit (SYBR Green, B532461, Sangon, Shanghai, China), following the manufacturer’s protocol. Next, the qRT-PCR reactions were performed and analyzed on the qTOWERE2.2 PCR machine (AnalytikJena, Germany). ΔCt value was used to show the expression level of an miRNA in one sample and ΔΔCt value was used to show the relative normalized expression (RNE, ) of an miRNA in two samples, as previously described (Chen et al. 2019).
Data availability
The small RNA sequencing data can be accessed from the NCBI Sequence Read Archive (SRA) platform (http://trace.ncbi.nlm.nih.gov/Traces/sra/) under the accession number PRJNA631631. All data supporting the results of this article are contained within the article and its Supplementary materials. Table S1: miRNAs identified in the Eucalyptus grandis genome. Table S2: miRNA expression profiles of the two Eucalyptus cultivars. Table S3: Differentially expressed miRNAs in the dedifferentiation of Eucalyptus. Table S4: Biological processes of gene ontology regulated by the differentially expressed miRNAs in E. camaldulensis and E. grandis x urophylla. Table S5: Primer sequences used for the tailed miRNA qRT-PCR. Supplementary material is available at figshare: https://doi.org/10.25387/g3.14151566.
Results
Global miRNA identification in Eucalyptus
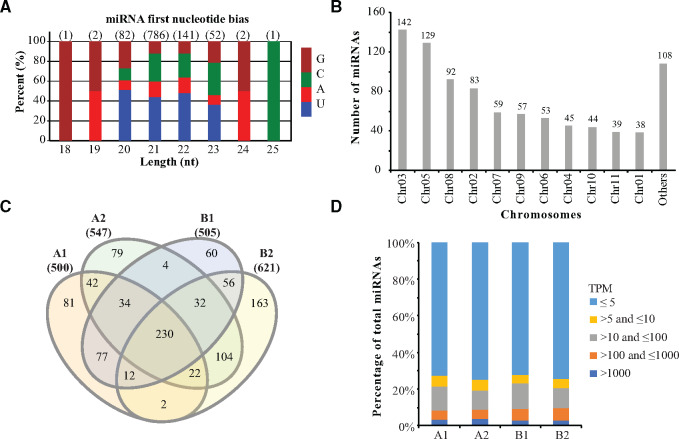
Due to the unavailable access of Eucalyptus miRNAs, we first identified miRNAs in the E. grandis genome using the small RNA sequencing data of differentiated (stem) and dedifferentiated (tissue culture–induced callus) of two Eucalyptus species (E. camaldulensis stem: A1; E. camaldulensis callus: A2; E. grandis x urophylla stem: B1; E. grandis x urophylla callus: B2). Initially, we obtained 346.4 million raw reads (average 28.9 million reads) and 304.2 million clean reads (average 25.4 million reads). Using the clean data, we identified 888 miRNA precursors (producing 1067 mature miRNAs, Supplementary Table S1), including 691 novel and 197 known miRNAs. Length distribution (Figure 1A) showed that 786 Eucalyptus mature miRNAs were in the length of 21 nt, which took 73.7% of the total miRNAs. Figure 1A also showed that the first nucleic acid of the miRNAs was enriched with uracil (U), which agrees with the base bias of plant miRNAs in miRBase (Griffiths-Jones et al. 2008). Next, we analyzed the chromosome distribution of the Eucalyptus miRNAs. Figure 1B showed that chromosome Chr03 can produce the most miRNAs (142 miRNA precursors), followed by chromosomes Chr05 (129 miRNA precursors) and Chr08 (92 miRNA precursors). Based on the miRNA locations in the Eucalyptus genome, we identified 46 miRNA clusters whose members are located within 10 kb (Supplementary Table S1) and have the potential of being transcribed together. We also found that 55, 52, and 782 miRNA precursors were located in the exon, intron, and intergenic regions, respectively. The redundant one was egd-MIR169aj, which can be produced by two chromosomes—Chr03 and Chr09 (Supplementary Table S1). These miRNAs were used as the reference for downstream analyses, including the miRNA expression profiling and target prediction.
Figure 1.
Overview of small RNA sequencing. (A) Length distribution of miRNAs identified in Eucalyptus. (B) Number of miRNAs identified in all chromosomes. (C) Comparison of miRNAs identified in the Eucalyptus camaldulensis (A1: differentiated, A2: dedifferentiated) and E. grandis x urophylla (B1: differentiated, B2: dedifferentiated). Only miRNAs with TPM > 5 were shown. (D) Expression levels of miRNAs identified in each sample. Mean values of miRNA expression in each sample (n = 3) were applied.
miRNA expression profiles
We next profiled the miRNA expression in the differentiated and dedifferentiated tissues of E. camaldulensis and E. grandis x urophylla. After lowly expressed miRNAs (average TPM < 5) were filtered, we identified 998 miRNAs (Figure 1C, Supplementary Table S2), of which 500, 547, 505, and 620 distributed in A1, A2, B1, and B2, respectively. Venn diagram (Figure 1C) showed that 230 miRNAs were detected in all samples, whereas 81, 79, 60, and 163 miRNAs were specifically identified in A1, A2, B1, and B2, respectively. The distribution of miRNAs in different expression levels revealed that more than 60% of the miRNAs were expressed <5 TPM (Figure 1D), whereas 31–38 miRNAs were expressed more than 1000 TPM in these samples. We showed in Table 1 the top 10 highly expressed miRNAs in these samples. It is interesting that the only the top three highly expressed miRNAs were shared by all four samples and that MIR166 family members were highly expressed in the dedifferentiated tissues of both E. camaldulensis and E. grandis x urophylla.
Table 1.
Top 10 highly expressed miRNAs identified in the differentiated and dedifferentiated tissues of E. camaldulensis and Eucalyptus grandis x urophylla
| Order | A1 | A2 | B1 | B2 |
|---|---|---|---|---|
| 1 | egd-miR398a-3p | egd-miR398a-3p | egd-miR398a-3p | egd-miR398a-3p |
| 2 | egd-miR398b-3p | egd-miR398b-3p | egd-miR398b-3p | egd-miR398b-3p |
| 3 | egd-N-miR1-3p | egd-N-miR1-3p | egd-N-miR1-3p | egd-N-miR1-3p |
| 4 | egd-miR166d-3p | egd-miR482a-3p | egd-miR166d-3p | egd-miR482a-3p |
| 5 | egd-miR166h-3p | egd-miR482a-5p | egd-miR166h-3p | egd-miR397a-5p |
| 6 | egd-N-miR2-3p | egd-miR397a-5p | egd-miR166c-3p | egd-miR397b-5p |
| 7 | egd-miR166f-3p | egd-miR397b-5p | egd-miR166f-3p | egd-miR159b-3p |
| 8 | egd-miR166g-3p | egd-N-miR2-3p | egd-miR166g-3p | egd-miR159a-3p |
| 9 | egd-miR166a-3p | egd-miR159b-3p | egd-miR166e-3p | egd-miR482a-5p |
| 10 | egd-miR166c-3p | egd-miR159a-3p | egd-miR166a-3p | egd-miR168a-5p |
Differentially expressed miRNAs in E. camaldulensis
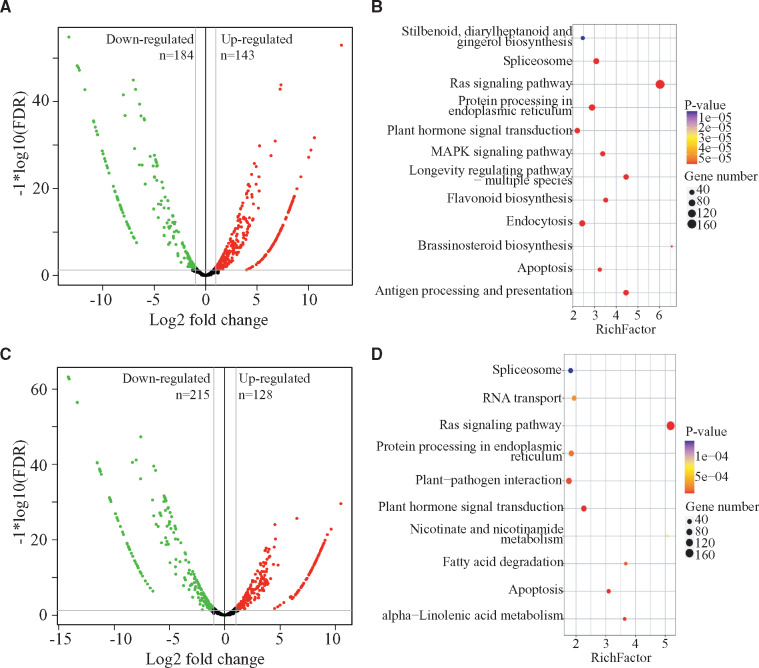
We next compared the miRNA profiles of differentiated and dedifferentiated tissues in E. camaldulensis. Compared with the differentiated tissue, we first identified 143 upregulated and 184 downregulated miRNAs in the dedifferentiated tissue of E. camaldulensis (Figure 2A, Supplementary Table S3). Among them, we found that 3 MIR167, 26 MIR169, 7 MIR3627, and 3 MIR159 members were upregulated and that 11 MIR156, 4 MIR164, 9 MIR166, 8 MIR171, 7 MIR395, and 43 MIR399 were downregulated. Target prediction showed that 303 of the DEmiRs can target 846 genes in E. camaldulensis (Table 2). For example, 130 upregulated miRNAs can target 405 genes including Eucgr.A00998 (transcription factor PIL1), Eucgr.B03766/Eucgr.G03183 (transcription factor GAMYB), Eucgr.G01528 (acetyl-CoA-benzylalcohol acetyltransferase), and Eucgr.B03050 (AP2/ERF and B3 domain-containing transcription factor RAV1); and 173 downregulated miRNAs in dedifferentiated tissue can target 373 genes including auxin response factors (e.g., Eucgr.D00264, Eucgr.F04380, Eucgr.G02838, Eucgr.J00923, Eucgr.K01240), growth-regulating factors (Eucgr.A01418, Eucgr.A01534, Eucgr.C00823, Eucgr.D01674, Eucgr.D02603, Eucgr.F00097, and Eucgr.F04420), Eucgr.F02691 (ethylene-responsive transcription factor ABR1) and Eucgr.K01046 (squamosa promoter-binding protein).
Figure 2.
Differentially expressed miRNAs identified in Eucalyptus camaldulensis and E. grandis x urophylla. (A) Volcano plot of upregulated (red) and downregulated (green) miRNA identified in the dedifferentiated tissue of E. camaldulensis compared with the differentiated tissue. (B) KEGG pathway enrichment by the target genes of differentially expressed miRNAs in E. camaldulensis. (C) Volcano plot of upregulated (red) and downregulated (green) miRNA identified in the dedifferentiated tissue of E. grandis x urophylla compared with the differentiated tissue. (D) KEGG pathway enrichment by the target genes of differentially expressed miRNAs in E. grandis x urophylla.
Table 2.
Target prediction of differentially expressed miRNAs in Eucalyptus camaldulensis and E. grandis x urophylla
| Cultivar | Type | miRNA | Targets | Examples |
|---|---|---|---|---|
| E. camaldulensis | Total | 310 | 1401 | |
| Upregulated | 135 | 819 | Transcription factor PIL1, transcription factor GAMYB, growth-regulating factor 1, AP2/ERF and B3 domain-containing transcription factor RAV1 | |
| Downregulated | 175 | 608 | Auxin response factors, transcription factor MYB1R1, ethylene-responsive transcription factor ABR1, translation initiation factor IF-2 | |
| E. grandis x urophylla | Total | 314 | 1550 | |
| Upregulated | 105 | 783 | Transcription factor PIL1, transcription factor bHLH69, probable WRKY transcription factor 19, growth-regulating factor 1, auxin response factor 6, transcription factor MYB23 | |
| Downregulated | 209 | 810 | Factor of DNA methylation 1, transcription factor bHLH95, transcription factor GAMYB, transcription factor IBH1, transcription factor MYB108, truncated transcription factor CAULIFLOWER A |
Next, we analyzed the potential pathways and biological processes involved by the differentially expressed miRNAs in the dedifferentiated tissue of E. camaldulensis. KEGG pathway enrichment (Figure 2B) showed that 160, 49, 45, 36, 9, and 3 target genes were involved in “Ras signaling pathway (ko04014),” “Plant hormone signal transduction (ko04075),” “Longevity regulation pathway—multiple species (ko04213),” “Flavonoid biosynthesis (ko00941),” “Zeatin biosynthesis (ko00908),” and “Aflatoxin biosynthesis (ko00254),” respectively. Gene ontology annotation (Supplementary Table S4) showed that the target genes of differentially expressed miRNAs in dedifferentiated tissue have the potential of regulating the biological processes such as “GO:0009809–lignin biosynthetic process,” “GO:0009744–response to sucrose,” “GO:0009909–regulation of flower development,” and “GO:0010075–regulation of meristem growth.”
Differentially expressed miRNAs in E. grandis x urophylla
In E. grandis x urophylla, we identified 128 upregulated and 215 downregulated miRNAs in dedifferentiation process (Figure 2C, Supplementary Table S3). The callus upregulated miRNAs include 13 MIR169, 4 MIR3627, 3 MIR390, and 4 MIR482, whereas the callus downregulated miRNAs include 21 MIR156, 3 MIR160, 11 MIR166, 44 MIR169, 12 MIR171, and 43 MIR399. We found that 314 DEmiRs in the dedifferentiated tissue of E. grandis x urophylla were predicted to target 1550 genes (Table 2). In details, 105 upregulated miRNAs target 783 genes, including Eucgr.A00998 (transcription factor PIL1), Eucgr.C00360 (transcription factor bHLH69), Eucgr.C00823 (growth-regulating factor 1), Eucgr.C01912 (probable WRKY transcription factor 19), Eucgr.D00264 (auxin response factor 6), and Eucgr.J02212 (transcription factor MYB23), and 209 downregulated miRNAs target 810 genes, such as auxin response factors (e.g., Eucgr.F04380, Eucgr.G02838, Eucgr.K01240, Eucgr.J00923, Eucgr.K03433, Eucgr.D00264), Eucgr.F03952 (factor of DNA methylation 1), Eucgr.H01993 (myb family transcription factor EFM), Eucgr.G01493 (transcription factor bHLH95), transcription factor GAMYB (Eucgr.B03766, Eucgr.G03183), Eucgr.K03562 (transcription factor MYB108), Eucgr.C00212 (transcription factor IBH1), and Eucgr.I02059 (truncated transcription factor CAULIFLOWER A). Functional analysis (Figure 2D) showed that the differentially expressed miRNAs in the callus tissue of E. grandis x urophylla were involved by pathways such as Aflatoxin biosynthesis (ko00254), “Brassinosteroid biosynthesis (ko00905),” “ABC transporters (ko02010),” “Spliceosome (ko03040),” and “Estrogen signaling pathway (ko04915).” GO analysis (Supplementary Table S4) of the target genes revealed that GO:0009809–lignin biosynthetic process, “GO:0006952–defense response,” “GO:0006355–regulation of transcription, DNA-templated,” “GO:0009658–chloroplast organization,” and “GO:0009867–jasmonic acid-mediated signaling pathway” were significantly enriched by the differentially expressed miRNAs in dedifferentiation process of E. grandis x urophylla. Based on the functional analysis, we found that the biological processes and pathways regulated by the differentially expressed miRNAs in E. camaldulensis and E. grandis x urophylla were different, which might indicate the different embryogenetic potentials of these two Eucalyptus cultivars.
miRNAs related to somatic embryogenesis
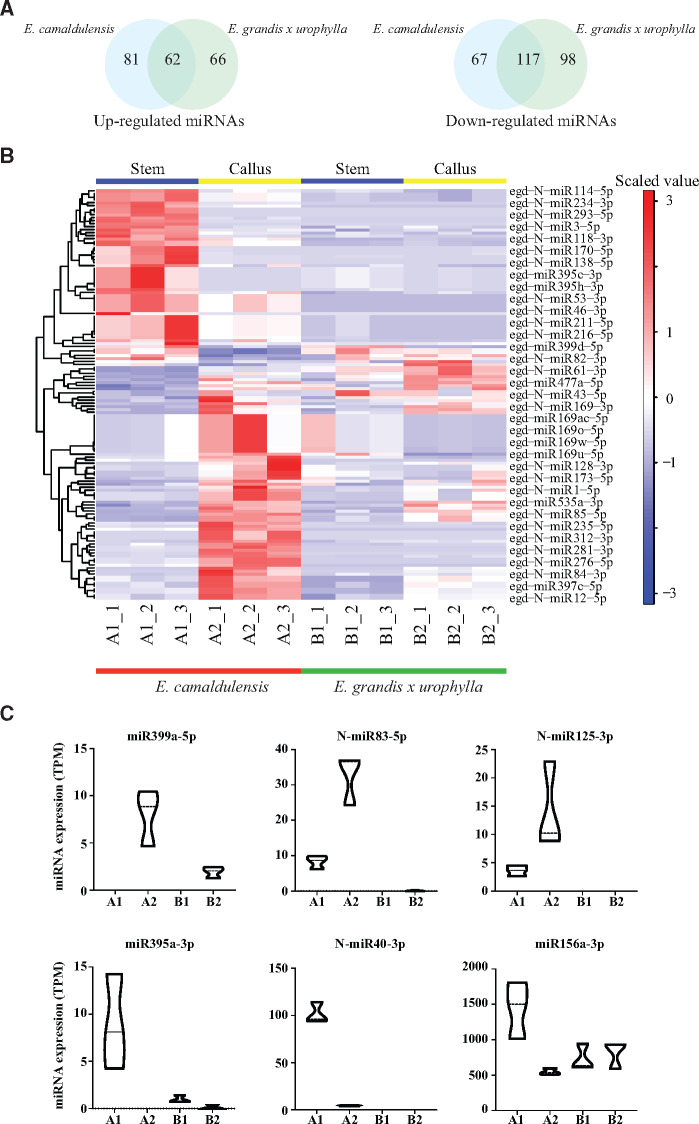
To investigate the miRNAs related to the embryogenetic potential of Eucalyptus, we next compared the DEmiRs in E. camaldulensis and E. grandis x urophylla. It showed that they shared 62 upregulated and 117 downregulated miRNAs (Figure 3A, Supplementary Table S3), including miRNAs from families MIR156, MIR159, MIR160, MIR164, MIR166, MIR169, MIR171, MIR399, and MIR482. We also identified 81 upregulated (e.g., miR159c-3p, miR167a-5p, miR397a-3p, miR397c-5p, miR397d-3p, miR397d-5p, N-miR1-5p, N-miR5-5p) and 67 downregulated (e.g., miR482b-3p, N-miR3-3p, miR156a-3p, N-miR40-3p, N-miR18-5p) miRNAs specific to E. camaldulensis (Figure 3A). The Heatmap (Figure 3B) confirmed the dysregulation of these miRNAs specific to E. camaldulensis and showed the expression patterns across the differentiated and dedifferentiated tissues of E. camaldulensis and E. grandis x urophylla. In Figure 3C, we listed the expression levels of 6 miRNAs (e.g., miR399a-5p, N-miR83-5p, N-miR125-3p, miR395a-3p, N-miR40-3p, miR156a-3p) specifically dysregulated in E. camaldulensis. Functional analysis of the targets of E. camaldulensis specifically dysregulated miRNAs revealed that they were involved in the pathways including “MAPK signaling pathway (ko04010),” Longevity regulating pathway—multiple species (ko04213), Estrogen signaling pathway (ko04915), Plant hormone signal transduction (ko04075), and Brassinosteroid biosynthesis (ko00905).
Figure 3.
miRNAs involved in the dedifferentiation of Eucalyptus callus. (A) Comparison of upregulated (left) and downregulated (right) miRNAs identified in the dedifferentiated tissue compared with the differentiated tissue in E. camaldulensis and E. grandis x urophylla. (B) Heatmap of differentially expressed miRNAs specifically identified in E. camaldulensis. Only some DEmiRs were shown in the plot by the program. (C) Box plots of the expression patterns for six miRNAs (miR399a-5p, N-miR83-5p, N-miR125-3p, miR395a-3p, N-miR40-3p, and miR156a-3p) in all samples.
Next, we analyzed the expression of SE-related DEmiRs and their targets (Table 3). Together with the transcriptome profiles of the same material, we observed the negative regulation of MIR160~ARF18, MIR396~GRF6, MIR1606~THB15/HD-ZIP, and MIR156/MIR157~SPL1. In addition, MIR171 and MIR535 was predicted to regulate the SCL6 (squamosa promoter-binding protein 1) and Eucgr.E01067 (glucan endo-1,3-beta-glucosidase 11)/Eucgr.K00316 (hypothetical protein), respectively. Interestingly, transcription factor (e.g., WRKY, MYB, GAMYB, TCP4, PIL1) and ribosomal protein genes were identified to be potential targets of DEmiRs in E. camaldulensis and/or E. grandis x urophylla (Table 3). Notably, although egd-N-miR63-5p and MIR169 were found to be upregulated in both E. camaldulensis and E. grandis x urophylla, their target genes PIL1 and RPS21C (40S ribosomal protein S21-2) were downregulated only in the dedifferentiated tissues compared with differentiated tissues of E. camaldulensis. Whether these miRNAs and their targets have functions in the SE and embryogenetic potential requires further experiments to be explored.
Table 3.
Expression changes of SE-related miRNAs and their target genes
| miRNA | Log2FC of miRNA |
Target | Log2FC of target gene |
Description | ||
|---|---|---|---|---|---|---|
| A2 vs A1 | B2 vs B1 | A2 vs A1 | B2 vs B1 | |||
| egd-miR160a-5p | −2.631 | −4.801 | Eucgr.K01240 | 0.526 | −0.501 | Auxin response factor 18 |
| egd-miR160b-5p | −2.631 | −4.802 | ||||
| egd-miR396a-5p | −3.661 | −3.236 | Eucgr.F00097 | 1.117 | −3.179 | Growth-regulating factor 6 |
| egd-miR396b-5p | −3.670 | −3.237 | ||||
| egd-miR396c-5p | −3.199 | −1.537 | ||||
| egd-miR396d-5p | −3.200 | −1.534 | ||||
| egd-miR166a-3p | −4.791 | −5.388 | Eucgr.F03066 | 1.585 | −0.282 | Homeobox-leucine zipper protein ATHB-15 |
| egd-miR166b-3p | −4.858 | −5.443 | ||||
| egd-miR166c-3p | −4.772 | −5.307 | ||||
| egd-miR166d-3p | −4.689 | −5.097 | ||||
| egd-miR166e-3p | −4.833 | −5.400 | ||||
| egd-miR166f-3p | −4.787 | −5.371 | ||||
| egd-miR166g-3p | −4.787 | −5.371 | ||||
| egd-miR166h-3p | −4.689 | −5.097 | ||||
| egd-miR166a-3p | −4.791 | −5.388 | Eucgr.D00184 | 5.012 | 2.379 | Homeobox-leucine zipper protein HOX32 |
| egd-miR166b-3p | −4.858 | −5.443 | ||||
| egd-miR166c-3p | −4.772 | −5.307 | ||||
| egd-miR166d-3p | −4.689 | −5.097 | ||||
| egd-miR166e-3p | −4.833 | −5.400 | ||||
| egd-miR166f-3p | −4.787 | −5.371 | ||||
| egd-miR166g-3p | −4.787 | −5.371 | ||||
| egd-miR166h-3p | −4.689 | −5.097 | ||||
| egd-miR166a-3p | −4.791 | −5.388 | Eucgr.B02504 | 2.994 | 2.755 | Homeobox-leucine zipper protein REVOLUTA |
| egd-miR166b-3p | −4.858 | −5.443 | ||||
| egd-miR166c-3p | −4.772 | −5.307 | ||||
| egd-miR166d-3p | −4.689 | −5.097 | ||||
| egd-miR166e-3p | −4.833 | −5.400 | ||||
| egd-miR166f-3p | −4.787 | −5.371 | ||||
| egd-miR166g-3p | −4.787 | −5.371 | ||||
| egd-miR166h-3p | −4.689 | −5.097 | ||||
| egd-miR166a-5p | −1.066 | −1.648 | Eucgr.B00437 | 4.417 | 1.582 | Probable WRKY transcription factor 61 |
| egd-miR171a-3p | −7.026 | −6.251 | Eucgr.E01510 | 1.156 | 1.610 | Scarecrow-like protein 6 |
| egd-miR171b-3p | −6.803 | −4.935 | ||||
| egd-miR171c-3p | −6.804 | −4.936 | ||||
| egd-miR171d-3p | −1.231 | −1.953 | ||||
| egd-miR171i-3p | −4.298 | −3.049 | ||||
| egd-miR171j-3p | −4.298 | −3.049 | ||||
| egd-miR156b-5p | −1.066 | −2.518 | Eucgr.K01046 | 2.361 | 2.611 | Squamosa promoter-binding protein 1 |
| egd-miR156l-5p | −3.745 | 0.726 | ||||
| egd-miR157a-5p | −5.944 | −5.412 | ||||
| egd-miR157b-5p | −5.981 | −5.475 | ||||
| egd-miR157c-5p | −5.944 | −5.412 | ||||
| egd-miR157d-5p | −5.944 | −5.412 | ||||
| egd-miR156b-5p | −1.066 | −2.518 | Eucgr.F02583 | 5.510 | 5.692 | Transcription repressor MYB6 |
| egd-miR156c-5p | −1.067 | −2.519 | ||||
| egd-miR535a-5p | −1.520 | −1.740 | Eucgr.E01067 | 1.001 | 2.267 | Glucan endo-1,3-beta-glucosidase 11 |
| egd-miR535b-5p | −1.525 | −1.745 | ||||
| egd-miR535a-5p | −1.520 | −1.740 | Eucgr.K00316 | 1.347 | 3.012 | Hypothetical protein |
| egd-miR535b-5p | −1.525 | −1.745 | ||||
| egd-N-miR145-3p | −4.962 | 0 | Eucgr.F02691 | 1.935 | −1.087 | Ethylene-responsive transcription factor ABR1 |
| egd-N-miR146-3p | −4.976 | 0 | ||||
| egd-miR159c-3p | 1.139 | −0.396 | Eucgr.B03766 | −1.461 | −2.900 | Transcription factor GAMYB |
| Eucgr.F01204 | −4.767 | −5.999 | Transcription factor TCP4 | |||
| egd-N-miR62-5p | 4.497 | 3.552 | Eucgr.A00998 | −4.333 | 0.165 | Transcription factor PIL1 |
| egd-N-miR63-5p | 4.497 | 3.552 | Eucgr.A00998 | −4.333 | 0.165 | Transcription factor PIL1 |
| egd-miR169ac-5p | 2.028 | −6.016 | Eucgr.B01917 | −1.333 | 0.517 | 40S ribosomal protein S21-2 |
| egd-miR169ad-5p | 1.972 | −6.016 | ||||
| egd-miR169ae-5p | 2.028 | −6.016 | ||||
| egd-miR169af-5p | 2.028 | −6.016 | ||||
| egd-miR169m-3p | 2.028 | −6.016 | ||||
| egd-miR169n-5p | 2.028 | −6.016 | ||||
| egd-miR169o-5p | 2.028 | −6.016 | ||||
| egd-miR169p-5p | 2.028 | −6.016 | ||||
| egd-miR169t-5p | 2.028 | −6.016 | ||||
| egd-miR169u-3p | 1.972 | −6.016 | ||||
| egd-miR169v-5p | 2.028 | −6.016 | ||||
| egd-miR169w-5p | 2.028 | −6.016 | ||||
| egd-miR169x-5p | 2.028 | −6.016 | ||||
Quantitative real time-PCR
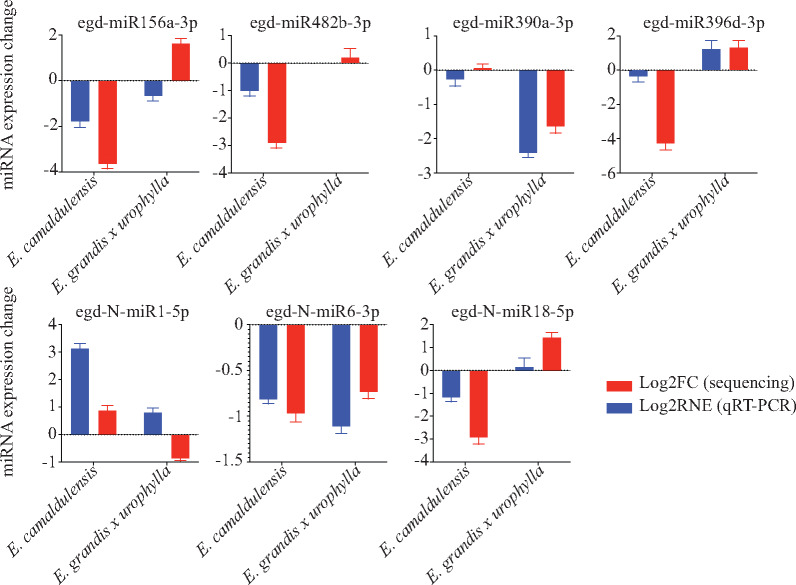
Next, we used tailed miRNA qRT-PCR strategy to validate the differential expression of seven randomly selected miRNAs in the differentiated and dedifferentiated tissues of E. camaldulensis and E. grandis x urophylla. The primer sequences for these miRNAs can be found in Supplementary Table S5. Comparison of small RNA sequencing and qRT-PCR results is shown in Figure 4. Overall, 12 (85.7%) out of 14 events were agreed by both small RNA sequencing and qRT-PCR. Using deep sequencing, egd-miR482b-3p, egd-miR156a-3p, and egd-N-miR1-5p were found with downregulation only in E. camaldulensis, whereas egd-miR390a-3p, egd-miR396d-3p, and egd-N-miR6-3p were found with dysregulation only in E. grandis x urophylla. The specific regulations of these miRNAs were confirmed by qRT-PCR. High agreement of miRNA expression patterns in small RNA sequencing and qRT-PCR indicate that the miRNAs identified in this study might be associated with the somatic embryogenesis and embryogenetic potential of Eucalyptus, which requires more experiments.
Figure 4.
qRT-PCR experiment. Log2FC and Log2RNE represent the log2 fold values of gene expression changes identified by small RNA sequencing and qRT-PCR, respectively.
Discussion
Although some studies have been demonstrated to study the Eucalyptus miRNAs (Levy et al. 2014; Pappas et al. 2015; Lin et al. 2018), the miRNA identities and sequences of Eucalyptus are still missing in miRbase (v22). In this study, we conducted the small RNA sequencing and identified a total of 888 miRNA precursors and 1067 mature miRNAs in the Eucalyptus genome (Supplementary Table S1). Among them, 197 precursors were aligned to other known miRNAs, whereas 691 could be specific to the Eucalyptus. The miRNAs identified in this study can provide a valuable source to study the regulation mechanisms of noncoding RNAs in the somatic embryogenesis of Eucalyptus. Comparison of miRNAs identified this study (Figure 1C) revealed 104 miRNAs specific to the callus tissues of Eucalyptus, including six highly expressed miRNAs (e.g., egd-miR482a-3p, egd-miR482a-5p, egd-miR397a-5p, egd-miR397b-5p, egd-miR159b-3p, egd-miR159a-3p) (Table 1). Although 79 miRNAs specifically identified in the callus tissue of E. camaldulensis (Figure 1C, Supplementary Table S2) might be related to the high potential of embryogenesis.
Among the top 10 highly expressed miRNAs (Table 1), miR482a, miR397a/b, and miR159a/b were shared by the dedifferentiated tissues of both E. camaldulensis and E. grandis x urophylla. In the callus-derived protoplast, both miR482a and miR482b were upregulated (Xu et al. 2017), and they were supposed to regulate the disease resistance pathways and the expression of plant protoplast totipotency (Wu et al. 2015). miR482 family has been proved to target mRNAs for nucleotide-binding site-leucine-rich repeats (NBS-LRR) disease resistance proteins using transient expression in Nicotiana benthamiana (Shivaprasad et al. 2012). In this study, no NBS-LRR gene was predicted to be regulated by miR482a, probably due to the limited annotation of Eucalyptus genes. However, we found that miR482a was predicted to target 30 genes encoding disease resistance proteins, such as Eucgr.A00735 (putative disease resistance protein At4g11170), Eucgr.A02363 (RGA1), Eucgr.C02142 (RPS6), Eucgr.H03707 (RML1B), Eucgr.H03723 (TAO1) and 14 genes encoding TMV resistance protein N. Interestingly, MIR397 has been reported to be particularly expressed in undifferentiated tissue and to be overexpressed in undifferentiated rice callus (Luo et al. 2006; Chen et al. 2011), which coincide with our results. MIR397 was reported to target a family of copper-containing oxidase enzymes—laccase, which is proposed to function in the formation of lignin, an integral part of the secondary cell walls of plants (Luo et al. 2006). We found a total of 112 laccase genes regulated by MIR397 in Eucalyptus. Also, miR397b-3p was upregulated in the callus of both E. camaldulensis and E. grandis x urophylla, and miR397a-3p was upregulated only in the dedifferentiated tissue of E. camaldulensis (Supplementary Table S3). Like MIR397, MIR159 has been studied to regulate the development of callus in multiple plants, such as Dimocarpus longan (Lin and Lai 2013; Xu et al. 2020), Moringa oleifera (Pirro et al. 2019), and Larix leptolepis (Zhang et al. 2010). miR159 has been shown to be induced by ABA and to regulate MYB33 during the seed germination of Arabidopsis (Reyes and Chua 2007). Although in the present study miR159 was not differentially expressed in the dedifferentiated tissue, its high expression indicates a potential role in the dedifferentiation and the callus development.
In addition, we identified 81 upregulated and 67 downregulated miRNAs specifically in the dedifferentiated tissue of E. camaldulensis compared with E. grandis x urophylla (Figure 3A). Among them, some miRNAs were detected with relative high expression in the stem or callus, such as miR156a-3p, miR167a-5p, miR482b-3p, miR3488-3p, and miR535a-3p (Supplementary Table S2). miR156a and miR482b were downregulated in the callus of E. camaldulensis (Supplementary Table S3). In Arabidopsis, the repression of miR156a by MYB33, a target of miR159, has the capacity of accelerate the vegetative development through its target SPL9 (Guo et al. 2017). In E. grandis and in E. brachyphylla, the expression of miR156 was shown to have no relationship with the rooting ability (Levy et al. 2014). Thus, the relationship between miR156 and somatic embryogenesis in E. grandis requires further experiments to be explored. The expression of miR167a was found to be upregulated during the development of rice callus and its target ARF8 was found to be downregulated (Chun et al. 2013). miR535-3p is the passenger miRNA of miR535, which was relevant from their expression (Supplementary Table S2). miR535-5p was downregulated in both E. camaldulensis and E. grandis x urophylla (Supplementary Table S3). In rice, SPL14, the target of miR535, remains at a low level during the vegetative growth and can strengthen the ability of disease resistance in plant (Agirre et al. 2008). miR3488 was identified due to its similarity to an animal miRNA. More experiments are required to characterize its presence in Eucalyptus and the association with the somatic embryogenesis.
We also identified a number of novel miRNAs for Eucalyptus (Supplementary Table S1) and some of them were highly expressed in the differentiated and dedifferentiated tissues, such as egd-N-miR1-3p, egd-N-miR2-3p, egd-N-miR4-3p, egd-N-miR5-5p, and egd-N-miR7-5p (Table 1, Supplementary Table S2). The abundance of these miRNAs support that they might be specific to the stem or the callus of Eucalyptus. Further, some of them were found to be differentially expressed in this study, such as egd-N-miR1-3p, egd-N-miR2-3p, and egd-N-miR5-5p (Supplementary Table S3). Among them, 99 Eucalyptus novel miRNAs were identified with dysregulation specific in E. camaldulensis, such as egd-N-miR3-3p and egd-N-miR-5p (Supplementary Table S3). Target prediction identified Eucgr.B01315 ((-)-germacrene D synthase) is the target of egd-N-miR3-3p, whereas egd-N-miR5-5p can regulate the expression of Eucgr.C03404 (myosin-17) and Eucgr.E03882 (microtubule-associated protein 70-5). Although it is not clear about their functions in the SE, our results provide a valuable resource for future studies in Eucalyptus.
Conclusion
In this study, we analyzed the small RNA profiles of differentiated and dedifferentiated tissues of two Eucalyptus cultivars—E. camaldulensis and E. grandis x urophylla. In total, we identified 691 novel and 197 known miRNAs for Eucalyptus, most of which distributed in chromosome Chr03 and Chr05. In the stem and callus, we detected 998 miRNAs with TPM > 5, of which 230 shared by all samples. Further, we identified 143 upregulated and 184 downregulated miRNAs in the dedifferentiation process of E. camaldulensis, and 128 upregulated and 215 downregulated miRNAs in E. grandis x urophylla. Comparison of the differentially expressed miRNAs in dedifferentiated tissues revealed 62 upregulated and 117 downregulated miRNAs shared by both E. camaldulensis and E. grandis x urophylla, which might be related to the SE of Eucalyptus. Further, the upregulation of 81 miRNAs and downregulation of 67 miRNAs were specifically identified in E. camaldulensis. These miRNAs were shown to function in the regulation of pathways like Longevity regulating pathway—multiple species and Plant hormone signal transduction. The expression of miRNAs was confirmed by qRT-PCR and their relationship with SE was agreed with previous studies in other plants. This is the first time to study the association of miRNAs and SE in Eucalyptus. The output of this study will provide a valuable resource for future studies in Eucalyptus. More importantly, our findings will benefit the molecular research in this field and the breeding program of Eucalyptus.
B.C., Z.Q., and J.L. conceived and designed the experiments; Y.Z., Y.X., and X.Z. performed the experiments; Z.Q., J.L., L.Z., and H.L. analyzed the data; Z.Q. and J.L. wrote the manuscript; B.C. revised the manuscript. All the authors have read and approved the final version of manuscript.
Funding
This work was financially supported by the Guangxi Science and Technology Program (GuiKeAD18281086), the Guangxi Forestry Science and Technology Project (2016[11], 2014[033]), the National Natural Science Foundation of China (31400522), and the Department of Human Resources and Social Security of Guangxi Zhuang Autonomous Region (GuiCaiSheHan2018112).
Conflicts of interest: The authors declare that there is no conflict of interest.
Acknowledgments
We acknowledge Mr Dajie Zhou for his involvement of the small RNA sequencing in BGI-Shenzhen.
Literature cited
- Agirre X, Jimenez-Velasco A, San Jose-Eneriz E, Garate L, Bandres E, et al. 2008. Down-regulation of hsa-mir-10a in chronic myeloid leukemia cd34+ cells increases usf2-mediated cell growth. Mol Cancer Res. 6:1830–1840. doi:10.1158/1541-7786.MCR-08-0167 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Snyder JA, Bartel DP.. 2007. Common functions for diverse small RNAs of land plants. Plant Cell. 19:1750–1769. doi:10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Liu Q, Zhang YC, Qu LH, Chen YQ, et al. 2011. Genome-wide discovery and analysis of microRNAs and other small RNAs from rice embryogenic callus. RNA Biol. 8:538–547. doi:10.4161/rna.8.3.15199. [DOI] [PubMed] [Google Scholar]
- Chen M, Xu R, Rai A, Suwakulsiri W, Izumikawa K, et al. 2019. Distinct shed microvesicle and exosome microRNA signatures reveal diagnostic markers for colorectal cancer. PLoS One. 14:e0210003.doi:10.1371/journal.pone.0210003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova TD, Alejandri-Ramirez ND.. 2014. Microrna expression and regulation during plant somatic embryogenesis. pp. 111-123 In: Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications: Transcriptional Regulation and Chromatin Remodelling in Plants, edited by Alvarez-Venegas R, De la Peña C and Casas-Mollano J. Cham, Switzerland: Springer International Publishing. [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, et al. 2010. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327:78–81. doi:10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- Durand-Cresswell R, Boulay M, Franclet A.. 1982. Vegetative propagation of Eucalyptus, pp.150-181 In: Tissue Culture in Forestry, edited by Bonga JM and Durzan DJ. Dordrecht, Netherlands: Springer Netherlan. [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ.. 2008. Mirbase: tools for microRNA genomics. Nucleic Acids Res. 36:D154–D158. Doi:10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Xu Y, Shi M, Lai Y, Wu X, et al. 2017. Repression of mir156 by mir159 regulates the timing of the juvenile-to-adult transition in Arabidopsis. Plant Cell. 29:1293–1304. doi:10.1105/tpc.16.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Sugimoto K, Iwase A.. 2013. Plant callus: Mechanisms of induction and repression. Plant Cell. 25:3159–3173. doi:10.1105/tpc.113.116053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Chen M, Greening DW, He W, Rai A, et al. 2014. Deep sequencing of RNA from three different extracellular vesicle (ev) subtypes released from the human lim1863 colon cancer cell line uncovers distinct miRNA-enrichment signatures. PLOS One. 9:e110314.doi:10.1371/journal.pone.0110314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Yarra R, Zhou L, Zhao Z, Cao H.. 2020. MiRNAs as key regulators via targeting the phytohormone signaling pathways during somatic embryogenesis of plants. 3 Biotech. 10:495.doi:10.1007/s13205-020-02487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J, Wang WG, Wang SH, Chen F.. 2013. The study on auxin-mir167-arf8 signal pathway during the growth and development process of rice callus. Journal of Sichuan University. 4:863-868. [Google Scholar]
- Kalvari I, Nawrocki EP, Ontiveros-Palacios N, Argasinska J, Lamkiewicz K, et al. 2021. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 49:D192–D200. doi:10.1093/nar/gkaa1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaravel M, Uma S, Backiyarani S, Saraswathi MS, Vaganan MM, et al. 2017. Differential proteome analysis during early somatic embryogenesis in Musa spp. AAA cv. Grand Naine. Plant Cell Rep. 36:163–178. doi:10.1007/s00299-016-2067-y. [DOI] [PubMed] [Google Scholar]
- Levy A, Szwerdszarf D, Abu-Abied M, Mordehaev I, Yaniv Y, et al. 2014. Profiling microRNAs in Eucalyptus Grandis reveals no mutual relationship between alterations in mir156 and mir172 expression and adventitious root induction during development. BMC Genomics. 15:524.doi:10.1186/1471-2164-15-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Liang Z, He S, Zeng Y, Jing Y, et al. 2017. Genome-wide identification of leaf abscission associated microRNAs in sugarcane (Saccharum officinarum L.). BMC Genomics. 18:754. doi:10.1186/s12864-017-4053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, et al. 2009. Soap2: an improved ultrafast tool for short read alignment. Bioinformatics. 25:1966–1967. doi:10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Lin YL, Lai ZX.. 2013. Evaluation of suitable reference genes for normalization of microRNA expression by real-time reverse transcription PCR analysis during longan somatic embryogenesis. Plant Physiol Biochem. 66:20–25. doi:10.1016/j.plaphy.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Lin Z, Li Q, Yin Q, Wang J, Zhang B, et al. 2018. Identification of novel miRNAs and their target genes in Eucalyptus Grandis. Tree Genetics Genomes. 14:60.doi:10.1007/s11295-018-1273-x. [Google Scholar]
- Liu W, Chen M, Bai L, Zhuang Z, Fan C, et al. 2017. Comprehensive transcriptomics and proteomics analyses of pollinated and parthenocarpic litchi (Litchi chinensis Sonn.) fruits during early development. Sci Rep. 7:5401.doi:10.1038/s41598-017-05724-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wang C, Shen X, Liang H, Wang Y, et al. 2019. Comparative transcriptome analysis highlights the hormone effects on somatic embryogenesis in catalpa Bungei. Plant Reprod. 32:141–151. doi:10.1007/s00497-018-0349-y. [DOI] [PubMed] [Google Scholar]
- Lopez-Ruiz BA, Juarez-Gonzalez VT, Chavez-Hernandez EC, Dinkova TD.. 2018. MicroRNA expression and regulation during maize somatic embryogenesis. Methods Mol Biol. 1815:397–410. doi:10.1007/978-1-4939-8594-4_28. [DOI] [PubMed] [Google Scholar]
- Luo YC, Zhou H, Li Y, Chen JY, Yang JH, et al. 2006. Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Lett. 580:5111–5116. doi:10.1016/j.febslet.2006.08.046. [DOI] [PubMed] [Google Scholar]
- Pappas MdCR, Pappas GJ, Grattapaglia D.. 2015. Genome-wide discovery and validation of Eucalyptus small RNAs reveals variable patterns of conservation and diversity across species of Myrtaceae. BMC Genomics. 16:1113.doi:10.1186/s12864-015-2322-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirro S, Matic I, Guidi A, Zanella L, Gismondi A, et al. 2019. Identification of microRNAs and relative target genes in Moringa oleifera leaf and callus. Sci Rep. 9:15145. doi:10.1038/s41598-019-51100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulianmackal AJ, Kareem AV, Durgaprasad K, Trivedi ZB, Prasad K.. 2014. Competence and regulatory interactions during regeneration in plants. Front Plant Sci. 5:142.doi:10.3389/fpls.2014.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua NH.. 2007. ABA induction of mir159 controls transcript levels of two MTB factors during Arabidopsis seed germination. Plant J. 49:592–606. doi:10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- Sabana AA, Rajesh MK, Antony G.. 2020. Dynamic changes in the expression pattern of miRNAs and associated target genes during coconut somatic embryogenesis. Planta. 251:79.doi:10.1007/s00425-020-03368-4. [DOI] [PubMed] [Google Scholar]
- Shi R, Chiang VL.. 2005. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 39:519–525. doi:10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, et al. 2012. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 24:859–874. doi:10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Ma X, Pan L, Miao J, Fu J, et al. 2017. Transcriptome analysis of Taxillusi chinensis (DC.) Danser seeds in response to water loss. PLoS One. 12:e0169177. doi:10.1371/journal.pone.0169177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ma YK, Chen T, Wang M, Wang XJ.. 2012. Psrobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 40:W22–W28. doi:10.1093/nar/gks554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XM, Kou SJ, Liu YL, Fang YN, Xu Q, et al. 2015. Genomewide analysis of small RNAs in nonembryogenic and embryogenic tissues of citrus: microRNA- and siRNA-mediated transcript cleavage involved in somatic embryogenesis. Plant Biotechnol J. 13:383–394. doi:10.1111/pbi.12317. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Li J, Zhang Y, Zhang X, Liu H, et al. 2020. Transcriptome analysis identifies genes involved in the somatic embryogenesis of Eucalyptus. BMC Genomics. 21:803.doi:10.1186/s12864-020-07214-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen X, Chen Y, Zhang Q, Su L, et al. 2020. Genome-wide identification of miRNAs and their targets during early somatic embryogenesis in Dimocarpus longan Lour. Sci Rep. 10:4626.doi:10.1038/s41598-020-60946-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Xu X, Zhou Y, Zeng S, Kong W.. 2017. Identification of protoplast-isolation responsive microRNAs in citrus reticulata blanco by high-throughput sequencing. PLoS One. 12:e0183524.doi:10.1371/journal.pone.0183524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QL, Su LY, Zhang ST, Xu XP, Chen XH, et al. 2020. Analyses of microrna166 gene structure, expression, and function during the early stage of somatic embryogenesis in Dimocarpus longan Lour. Plant Physiol Biochem. 147:205–214. doi:10.1016/j.plaphy.2019.12.014. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhou J, Han S, Yang W, Li W, et al. 2010. Four abiotic stress-induced miRNA families differentially regulated in the embryogenic and non-embryogenic callus tissues of Larix leptolepis. Biochem Biophys Res Commun. 398:355–360. doi:10.1016/j.bbrc.2010.06.056. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhao H, Li W, Wu J, Zhou Z, et al. 2019. Genome-wide association study of callus induction variation to explore the callus formation mechanism of rice. J Integr Plant Biol. 61:1134–1150. doi:10.1111/jipb.12759. [DOI] [PubMed] [Google Scholar]
- Zimmerman JL. 1993. Somatic embryogenesis: a model for early development in higher plants. Plant Cell. 5:1411–1423. doi:10.2307/3869792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The small RNA sequencing data can be accessed from the NCBI Sequence Read Archive (SRA) platform (http://trace.ncbi.nlm.nih.gov/Traces/sra/) under the accession number PRJNA631631. All data supporting the results of this article are contained within the article and its Supplementary materials. Table S1: miRNAs identified in the Eucalyptus grandis genome. Table S2: miRNA expression profiles of the two Eucalyptus cultivars. Table S3: Differentially expressed miRNAs in the dedifferentiation of Eucalyptus. Table S4: Biological processes of gene ontology regulated by the differentially expressed miRNAs in E. camaldulensis and E. grandis x urophylla. Table S5: Primer sequences used for the tailed miRNA qRT-PCR. Supplementary material is available at figshare: https://doi.org/10.25387/g3.14151566.