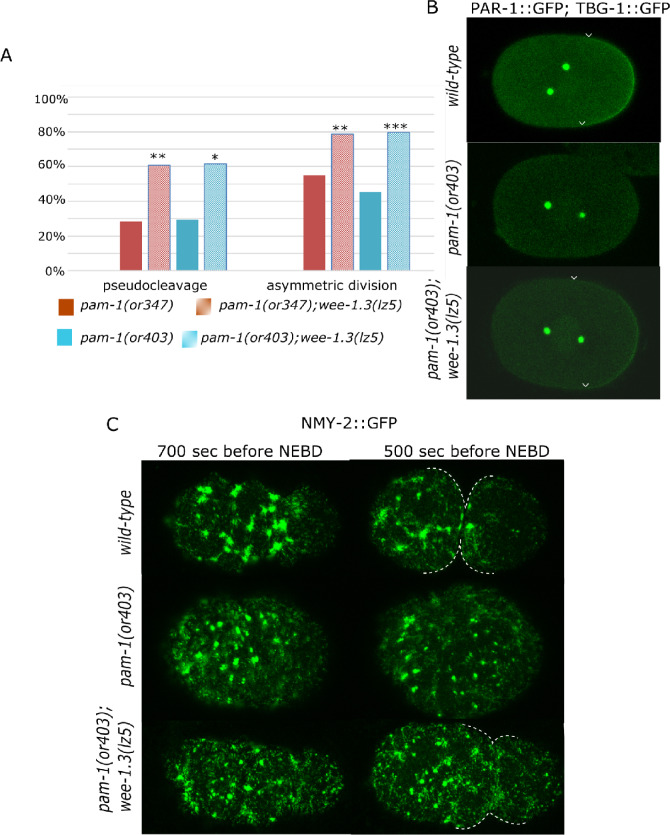
Figure 3.
Polarity is improved in pam-1; wee-1.3(lz5) embryos. (A) wee-1.3(lz5) suppresses polarity defects of two pam-1 alleles. The presence of pseudocleavage in pam-1(or347) (n = 39) and pam-1(or403) (n = 27) is increased in pam-1(or347); wee-1.3(lz5) (n = 46) and pam-1(or403); wee-1.3(lz5) (n = 34). Similarly, while many pam-1(or347) (n = 78) and pam-1(or403) (n = 46) embryos divide symmetrically, the presence of an asymmetric first cleaveage is improved in pam-1(or347); wee-1.3(lz5) (n = 85) and pam-1(or403); wee-1.3(lz5) (n = 54) embryos. (B) PAR-1 and gamma-tubulin GFP confocal images. PAR-1 localizes to the posterior in wild-type during polarization. Arrowheads mark the edges of the PAR-1 domain. Although many pam-1(or403) embryos lack posterior PAR-1 localization (Saturno et al. 2017), 100% of pam-1(or403); wee-1.3(lz5) embryos have posterior localization of PAR-1 in a domain size similar to wild-type (n = 46). C) NMY-2::GFP images show clearing from the posterior in wild-type, 700 seconds prior to NEBD and full pseudocleavage 200 seconds later (n = 18). Both pam-1(or403) (n = 18) and pam-1(or403); wee-1.3(lz5) (n = 13) embryos often lack posterior clearing. Shown here is reduced clearing in both strains, but pseudocleavage is restored in the presence of the suppressor. Pseudocleavage is marked by dotted lines. Anterior to the left in all panels. *P < 0.05; **P < 0.01; ***P < 0.001