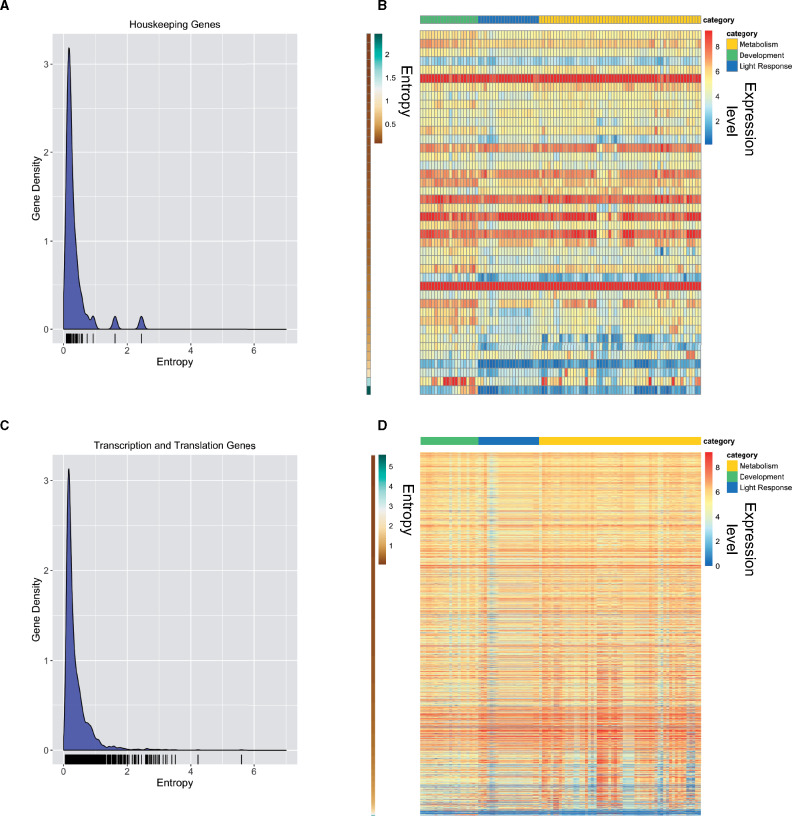
Figure 2.
Constitutively expressed genes are characterized by low entropy values. (A) The relative frequency of entropy values for a list of housekeeping genes is shown as a KDE plot. The rug plot, black lines on the bottom in the KDE plot, represents the individual data points that create the estimation. The y-axis is the probability density, which is the probability for each unit (gene) on the x-axis. The total area below the KDE curve integrates to one. (B) The heatmap shows the expression value for housekeeping genes across all conditions analyzed. The expression level for each gene is plotted as the log2 transformed TPM value. Genes (rows) are plotted in ranked order based on the entropy value from low (top) to high (bottom). The scale on the left indicates entropy values for each gene. Each condition (column) has been assigned a category: Metabolism (gold), Development (green), or Light Response (blue). The categories are represented at the top of the heatmap in the three different colors. (C) The relative frequency of entropy values for a list of genes related to transcription and translation is shown as a KDE plot. The rug plot, black lines on the bottom in the KDE plot, represents the individual data points that create the estimation. The y-axis is the probability density, which is the probability for each unit (gene) on the x-axis. (D) Heatmap of log2 transformed TPM values from all transcription and translation related genes (rows) ranked by entropy (low to high). Entropy values are depicted by the brown to green heatmap on the left, where brown is low (top) and green is high (bottom). Each condition (column) has been assigned a category: Metabolism (gold), Development (green), or Light Response (blue). The categories are represented at the top of the heatmap in the three different colors.