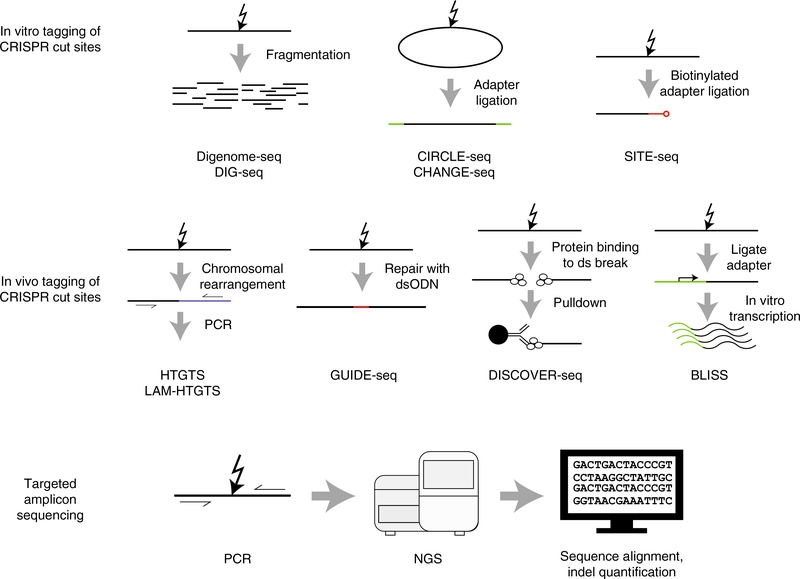
Fig. 2 |. Schematics showing in vitro and in vivo experimental techniques most commonly used to characterize off-target cutting by CRISPR–Cas9.
These techniques include those for detection of (i) in vitro (biochemical) tagging of Cas9 cut sites, (ii) in vivo (cellular) tagging of Cas9 cut sites and (iii) targeted sequencing of PCR amplicons. Cas9 binding techniques are not commonly used to characterize off-target cutting and are, therefore, not shown. Targeted amplicon sequencing is routinely used to verify off-target sites identified by these experimental techniques.