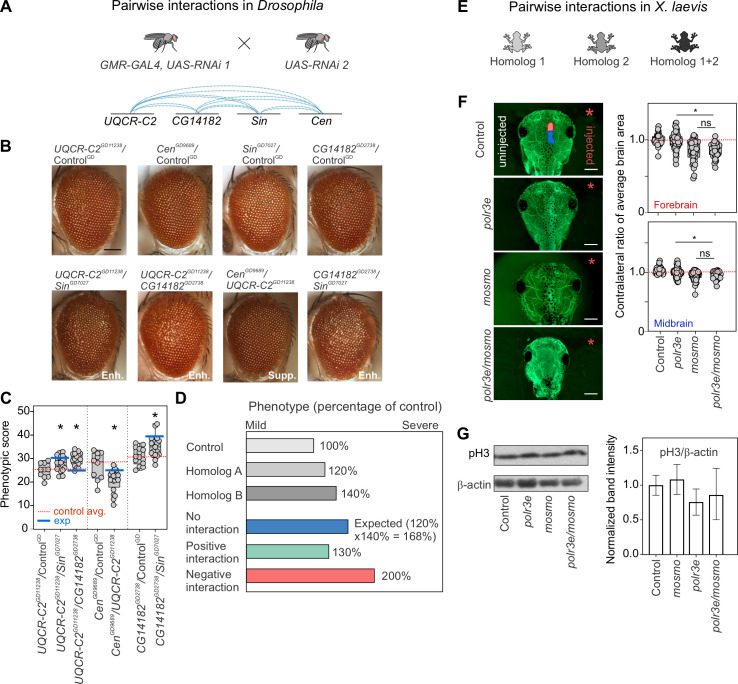
Fig 3. Homologs of 16p12.1 genes contribute towards neurodevelopmental defects through weak genetic interactions and combined independent effects.
(A) We generated eye-specific GMR-GAL4 recombinant lines for the four 16p12.1 homologs to test a total of twelve pairwise interactions for modulation of eye defects. (B) Representative brightfield images of Drosophila adult eyes for recombinant lines of 16p12.1 homologs crossed with RNAi lines for the other homologs, which show enhancement (Enh.) or suppression (Supp.) of the phenotypes observed with crosses with control. Scale bar represents 100 μm. (C) Simultaneous knockdown of UQCR-C2GD11238 with CG14182GD2738 (n = 18, two-tailed Mann-Whitney with Benjamini-Hochberg correction, *p = 0.002) or SinGD7027 (n = 19, *p = 0.023) led to a significant enhancement in the eye phenotype (measured using Flynotyper scores) compared to knockdown of UQCR-C2GD11238 alone. Similarly, simultaneous knockdown of CG14182GD2738 with SinGD7027 (n = 19, *p = 0.021) enhanced the eye phenotype observed for CG14182GD2738 alone. Simultaneous knockdown of CenGD9689 with UQCR-C2GD11238 (n = 20, *p = 0.023) led to a milder suppression of the eye phenotype compared to knockdown of CenGD9689 alone. Double knockdowns were compared to the recombinant lines of the 16p12.1 homologs crossed with wild-type controls for the second 16p12.1 homolog. Note that only experiments with ControlGD are represented here; see S8 Fig for results from other lines with KK and BL controls. (D) We applied a multiplicative model to identify the nature of combinatorial effects for the pairwise knockdowns tested. The expected phenotype from simultaneous knockdown of homolog A and homolog B, or when the combined effect indicates no genetic interaction (in blue), was calculated as the product of the normalized phenotypic scores (i.e. percentage of control) observed from knockdown of individual genes. Positive or alleviating genetic interactions were identified for combinations where the observed phenotype was significantly milder than expected (in green), while negative or aggravating interactions were identified when the combined phenotypes were significantly more severe than expected (in red). One-sample Wilcoxon signed rank tests with Benjamini-Hochberg correction for multiple testing were used to identify significant interactions. (E) We generated double knockdowns of 16p12.1 homologs in X. laevis models by co-injecting embryos with morpholinos of two homologs. All double knockdown experiments were performed with partial knockdown of the genes, to avoid potential lethality with stronger knockdown. (F) Representative images of tadpoles stained with anti-tubulin show forebrain (red on control image) and midbrain (blue) areas of the side injected with morpholino (right, red asterisk), which were normalized to the uninjected side (left). Simultaneous knockdown of polr3e and mosmo led to decreased forebrain (n = 36, two-tailed student’s t-test, *p = 1.10×10−9) and midbrain area (*p = 1.98×10−7), which showed no differences compared to partial knockdown of mosmo alone. Control data represents control injected with highest amount of morpholino (22ng). Scale bar represents 500 μm. (G) Representative western blots show bands for phosphorylated histone-3 (pH3) and β-actin for the uninjected control, knockdown of polr3e, knockdown of mosmo, and pairwise knockdown of polr3e and mosmo (full western blots are shown in S6 Fig). Bar plot shows intensity of pH3 band normalized to β-actin, with error bars representing mean ± SD. Simultaneous knockdown of polr3e and mosmo does not lead to changes in the proliferation defects observed with knockdown with polr3e alone. Boxplots represent all data points with median, 25th and 75th percentiles, and red dotted lines indicate the control median. Statistical details, including sample size, confidence intervals, and p-values, are provided in S6 File. A list of full genotypes for fly crosses used in these experiments is provided in S1 File.