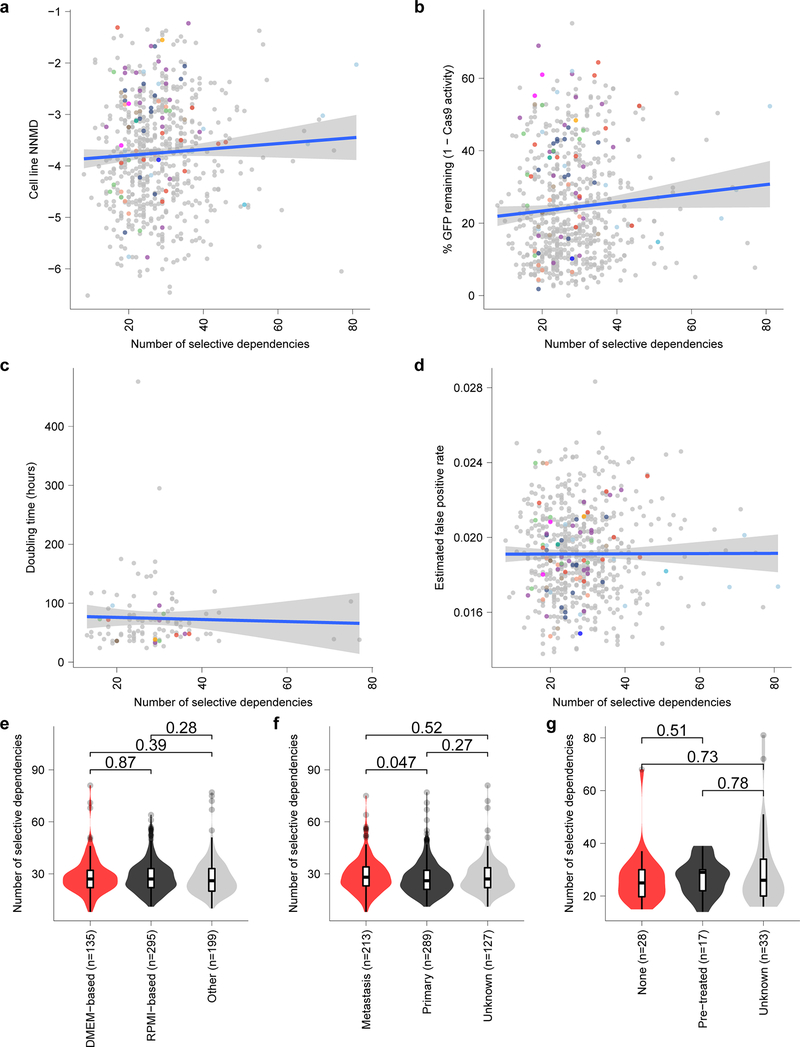
Extended Data Fig. 5. Selective dependencies in pediatric cell lines and the relationship to confounders.
a, Screen quality measured by null-normalized mean difference (NNMD) between positive and negative controls (y-axis) compared to number of selective dependencies per cell line (x-axis). b, Cas9 activity expressed as percent of GFP remaining after CRISPR-Cas9-mediated disruption of exogenous GFP (y-axis) compared to number of selective dependencies per cell line (x-axis). c, Cell line doubling time (y-axis) compared to number of selective dependencies per cell line (x-axis). d, Estimated false positive rate calculated as the fraction of genetic dependencies in a cell line that are not expressed in RNA sequencing data (y-axis) compared to the number of selective dependencies per cell line (x-axis). Circles in panels (a-d) represent individual cell lines with tumor types colored as in panel (Extended Data Fig. 4e). Blue lines in panels (a-d) represent a linear model fit to this data with gray shaded area representing the 95% confidence interval around the fit. e, Number of selective dependencies in cell lines cultured in DMEM-based media (red, n=135 biologically independent cell lines), RPMI-based media (black, n=295 biologically independent cell lines), or other media (gray, n=199 biologically independent cell lines). f, Number of selective dependencies per cell line annotated as derived from metastatic samples (red, n=213 biologically independent cell lines), primary tumors (black, n=289 biologically independent cell lines), or unknown (gray, n=127 biologically independent cell lines). g, Number of selective dependencies per pediatric cancer cell line annotated by literature search as derived from a patient with no pre-treatment (“none”, red, n=28 biologically independent cell lines), after treatment (“pre-treated”, black, n=17 biologically independent cell lines), or unknown (gray, n=33 biologically independent cell lines). Horizontal lines in panels (e-g) demonstrate the median (center) with minima and maxima box boundaries demonstrating the 25 and 75th percentiles. Upper and lower bounds (whiskers) in panels (e-g) represent 10 and 90th percentiles respectively.