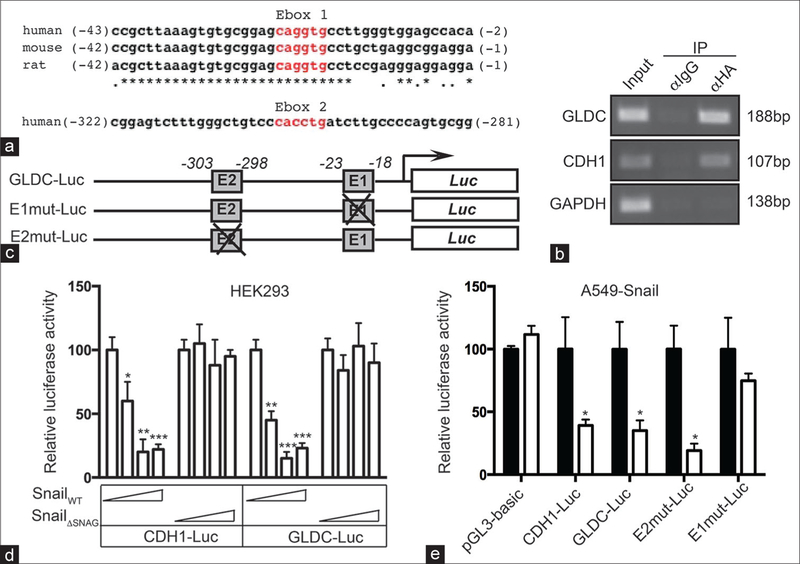
Figure 5:
Snail suppresses the promoter of glycine decarboxylase gene by binding to an evolutionarily conserved proximal E‑box motif. (a) glycine decarboxylase genomic sequences at the nucleotide positions from −43 to −2 (top, aligned with the indicated rodent sequences) and from −322 to −281 are exhibited. The E‑box motifs are highlighted in red. (b) A549‑Snail cells were grown in the presence of 2 μg/ml Dox for 2 days. The binding of Snail‑HA to the promoters of glycine decarboxylase, CDH1, and GAPDH were measured using chromatin immunoprecipitation with the anti‑HA antibody followed by polymerase chain reaction amplification with the gene‑specific primers. The amplified products were visualized using agarose gel electrophoresis, and the sizes of each individual amplicon are labeled at the right. (c) A schematic representation of the luciferase reporter constructs. (d) HEK293 cells were transfected with the indicated plasmids for 2 days, and the luciferase activities of the cells were measured. *P < 0.05, **P < 0.01, ***P < 0.001 (t‑test). (e) A549‑Snail cells were transfected with the indicated reporter constructs and then grown in the absence (black bars) and presence (white bars) of 2 μg/ml Dox for 2 days, and the luciferase activities of the cells were subsequently measured. *P < 0.05 (t‑test)