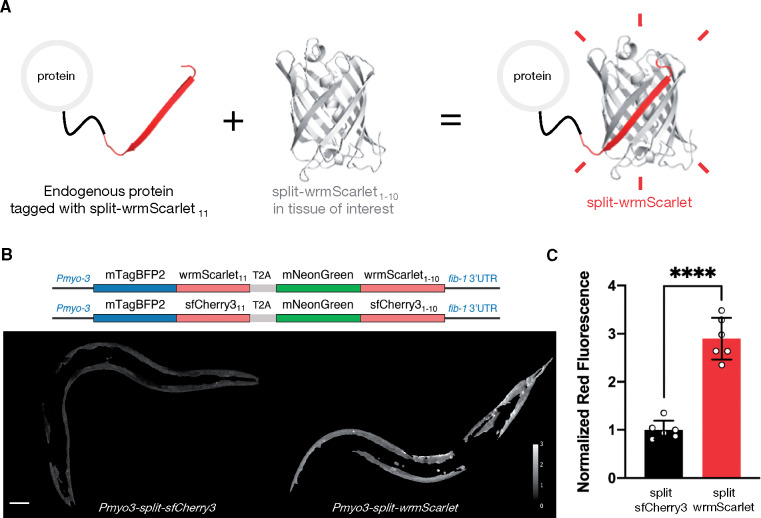
Figure 1.
Engineering and evaluating split-wrmScarlet. (A) Principle of endogenous protein labeling with split-wrmScarlet. The protein structure from split-wrmScarlet was generated using Phyre2 and PyMOL. (B) Schematic of the plasmids encoding split-wrmScarlet and split-sfCherry3. Each plasmid consists of the large FP1–10 sequence fused to mNeonGreen, and the corresponding small FP11 sequence fused to mTagBFP2. The T2A sequence ensures that mTagBFP2::FP11 and the corresponding mNeonGreen::FP1–10 are separated. The images are representative displays of the ratio of red to green fluorescence intensity from images acquired under identical conditions after background subtraction and masking with the same threshold. Scale bar, 50 µm. (C) Emission intensities from split-sfCherry3 and split-wrmScarlet normalized to mNeonGreen. Mean ± SD. Circles are individuals (n = 6 for each split fluorescent protein). ****P < 0.0001.