Figure 1. Fusion of mouse ESCs with human EBV-B lymphocytes, induces genome-wide transcriptional changes in the human nuclei.
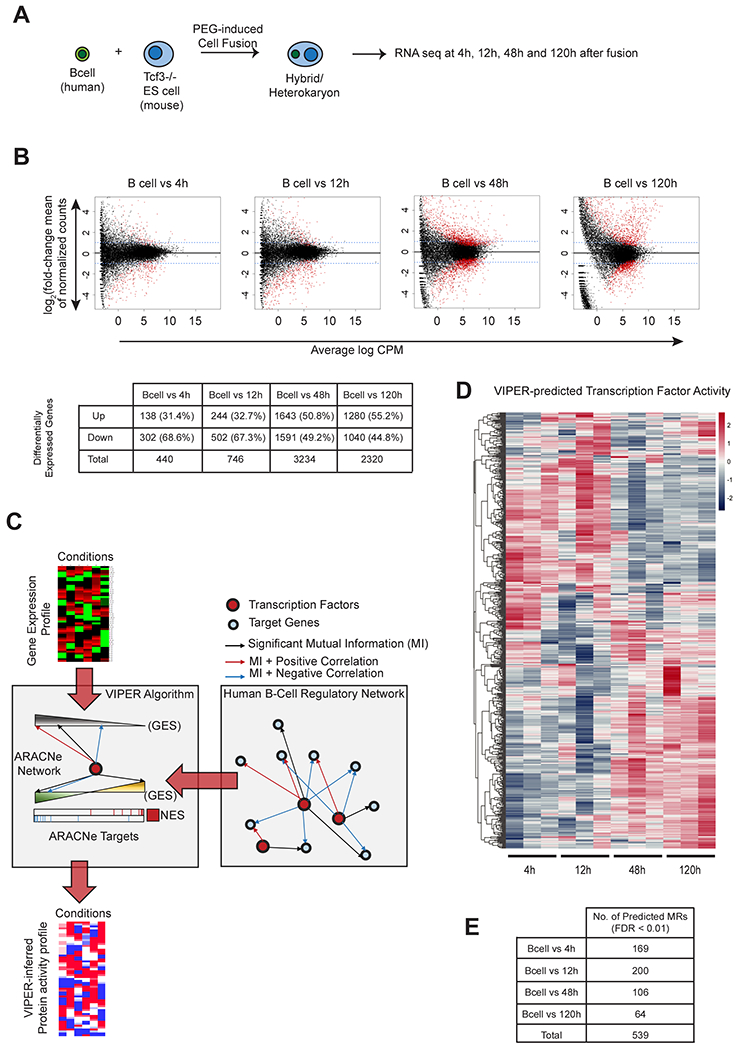
(A) Schematic for generation of heterokaryons and paired-end sequencing. (B) Differential expression of human genes after fusion of murine Tcf7l1−/− ESCs with human EBV-B lymphocytes. x-axis : Log2 fold-change of normalized counts and y-axis : mean of normalized counts. Blue-dashed line indicates fold-change of 1. Genes with significant (FDR < 0.05) change in expression are shown in red. Average log CPM (Count Per Million) values were calculated from 3 biological replicates for each time-point. Table shows upregulated and downregulated genes. See also Figure S1. (C) Schematic for the generation of protein activity profile using the VIPER algorithm and ARACNe network (See STAR Methods for details). (D) Heatmap of VIPER-predicted Normalized Enrichment Score values (NES values) of 539 MRs that had significant predicted activities (FDR < 0.01) in the heterokaryon samples. NES values were calculated using the VIPER algorithm by comparing each heterokaryon sample against the unfused PEG-treated B-cells. Positive NES values indicating active MRs are shown in red, and negative NES values indicating silenced MRs are shown in blue. (E) Number of MRs predicted in each timepoint (FDR < 0.01).
