Figure 5.
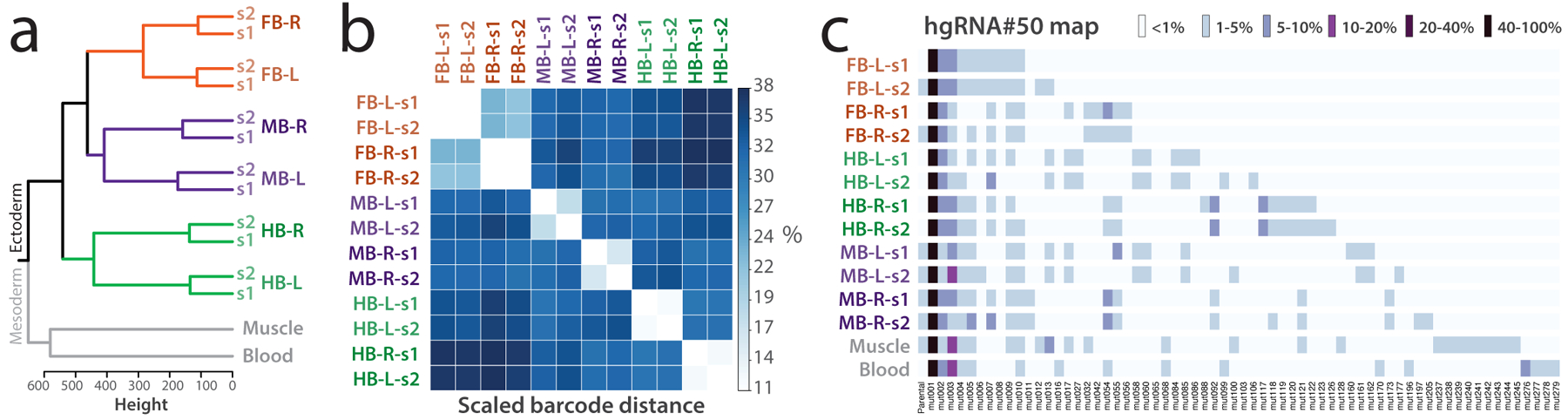
Example experimental design and results. (a) Outgroup selection and replicates provide internal controls. Clustering barcodes from 30 active hgRNAs in a twelve-month-old female mouse show the order of commitment to brain axes by neuronal progenitors. Mesoderm outgroups, in gray, clustered separately from ectodermal neuronal samples; duplicate samples (s1, s2) consistently clustered together, confirming consistency of barcoding across the board. Hierarchical clustering was carried out based on Manhattan distance of each sample’s full barcode (Supplementary Table 8) using Ward’s minimum variance method with squared dissimilarities (ward.D2)7. FB: Forebrain; MB: Midbrain; HB: Hindbrain; R: Right; L: Left. (b) Heatmap of the scaled pairwise barcode distances between all samples shown in a. The barcode distance is scaled such that a value of 0% corresponds to the presence of the same alleles at the exact same frequencies and a value of 100% corresponds to entirely non-overlapping sets of alleles. (c) Visual representation of a part of the barcode table showing the allele composition of one of the 30 active hgRNAs (hgRNA#50, ID: GTACACAATT) that contribute to the clustering in a. Only the 65 mutant alleles with 1% or more abundance in at least one sample are shown (among 302 in total). Full barcodes for all samples can be found in Supplementary Table 8. Panel a is reproduced with permission from ref. 7.
