Abstract
The emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) late last year has not only led to the world-wide coronavirus disease 2019 (COVID-19) pandemic but also a deluge of biomedical literature. Following the release of the COVID-19 open research dataset (CORD-19) comprising over 200,000 scholarly articles, we a multi-disciplinary team of data scientists, clinicians, medical researchers and software engineers developed an innovative natural language processing (NLP) platform that combines an advanced search engine with a biomedical named entity recognition extraction package. In particular, the platform was developed to extract information relating to clinical risk factors for COVID-19 by presenting the results in a cluster format to support knowledge discovery. Here we describe the principles behind the development, the model and the results we obtained.
Keywords: COVID-19, Text Mining, Cluster Algorithms, Biomedical NLP
Introduction
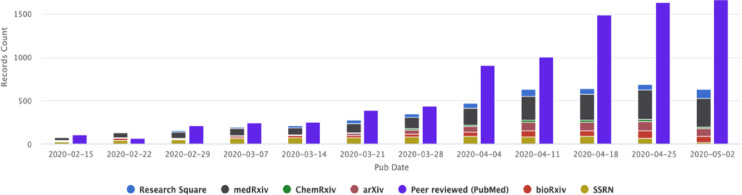
The first coronavirus disease 2019 (COVID-19) cases emerged in Wuhan city in China as a result of infection from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).1 Since then the disease has spread across the world resulting in a pandemic.2 To contain the disease, there is a world-wide effort to study the virus and devise treatments and vaccine candidates.3 Clinicians and researchers are in urgent need of rapid and quality information that will help them to inform diagnostics and therapeutics relating to the disease. However, the volume of studies concerning COVID-19 has substantially increased since the beginning of 2020 (fig. 1 ).4 When you couple these studies with pre-existing studies relating to other coronaviruses, immunology, genomics, proteomics and therapeutics the volume increases markedly.5
Fig. 1.
Increase in pre-print and peer reviewed publications relating to COVID-19 February-May 2020 (Source: NIH, 2020).
While not everything in this collection of medical literature is of proven value, it is nevertheless an essential source of information in the pandemic context.5 In this context, researchers and clinicians require a reliable approach to mining published literature for novel insights, emerging risk factors and therapeutics to inform their work in combating the COVID-19 pandemic. Biomedical natural language processing (NLP) or text mining can help mine useful information and knowledge from large volumes of biomedical literature.6 The objective of text mining is to derive implicit knowledge that hides in such literature and present it in an explicit form. In this context, we offer an innovative text mining and analytical tool that will aid clinicians and researchers in extracting valuable insights from large datasets of literature. In the following sections, we describe the principles, techniques and results of our approach.
Methods
The first step in our text mining process was information retrieval (IR). The dataset we relied on to retrieve information in this step was the COVID-19 Open Research Dataset (CORD-19), which is a resource of over 60,000 scholarly articles, including over 47,000 publications covering subjects such as COVID-19 and coronavirus related research.7 The CORD-19 dataset was initially made available for machine learning practitioners as an open-access database by a coalition including the White House, National Institutes of Health, and other partners but is now maintained by the Allen Institute for AI. The dataset gets updated weekly, and we last retrieved the dataset, to assess the utility of our model, in the first week of January 2021.
As part of the pre-processing seven sections in the full text of the publications were indexed for information retrieval including 'Abstract', 'Introduction', 'Background', 'Discussion', 'Results', 'Results and Discussion' and 'Methods'. The sections were established through an internal consultation process with the medical researchers in the team. Solr, an open-source enterprise search platform, was employed as a foundational query search engine for information extraction. Solr, uses okapi BM25 a ranking function used by search engines and provides distributed indexing and load-balanced querying, which suited our objectives.8
The input for the search engine was the CORD-19 dataset, with files in JavaScript Object Notation Format. The search engine allowed both free text and Boolean queries, while allowing unlimited queries. Written into the code was a process to eliminate duplicates of the retrieved information like for example similar abstracts. Weighted queries ensured only the most relevant articles were retrieved. The search platform was enjoined with scispaCy, a package tailored to identify within the search results biomedical or clinical terms.9 scispaCy builds on the Python-based spaCy library, which has a number of tools to aid text processing in multiple languages. scispaCY customises these tools to support primary text processing requirements for the biomedical, scientific and clinical areas. Initially, key terms relating to COVID-19 including all its associated synonyms were built into the search process to filter results. The search process then incorporated the scispaCy model to filter the results further and extract the medical terms.
Knowledge about the relations between biomedical factors are embedded in literature and extracting information about the relationships can be useful for medical research.10 Following retrieval of results from the search process, there is a requisite to obtain clarity from the results, which necessitated us to explore various analytical approaches. It has been identified that clustering is a useful analytical approach for gaining insights from biomedical data mining as it enables dimensionality reduction and visualisation of similarity indices. Clustering is a core task in many machine learning and text mining approaches.11 Clustering algorithms are known to be useful to explore large scale data.12 Cluster analysis when applied provides insights regarding underlying data similarity structures. The clustering technique groups similar terms into the same cluster thus assisting with knowledge discovery.
For the purpose of this project, we used spectral clustering (SC), which is stated to achieve better results than generative models.13 For dimensionality reduction and to organise retrieved results into clusters, we initially employed spectral embedding (SE). SE is generally employed for non-linear dimensionality reduction and works by forming an affinity matrix and applying spectral decomposition to the graph Laplacian. KNN algorithm was used to create heat kernels, which served as input for SE. SC uses the top eigenvectors of a matrix that is resulting from the distance between points. SC conserves linguistically motivated similarities. Also SC identifies communities of nodes in the dataset based on the edges connecting them.14 Unlike traditional approaches, SC utilises proximity or connectivity to cluster data instead of distance or compactness. SC represents data as graphs where samples are vertices and the similarity between samples is represented as edge weights.11 SC adopts an approach where given a data set X = {xi}n i=1, the purpose of SC is to separate X into c clusters. The cluster assignment matrix is represented by Y = [y1, y2, ..., yn]T ∈ B nXc, where yi ∈ B cX1 (1 ≤ i ≤ n) is the assigned vector for the pattern xi. Following this, the j-th element of yi is represented as 1 and if the pattern xi is assigned to the j-th cluster it is 0. Thus, the main task of a clustering algorithm is to learn the cluster assignment matrix Y.
To enable a more comprehensive approach and a comparison of clustering approaches we also utilised agglomerative clustering (AC) in our project. AC or hierarchical clustering as it is also known is another technique used for undertaking exploratory data analysis.15 AC commences with a large number of small clusters and through an iterative process selects two clusters with the greatest affinity until a stopping condition is attained.16 In other words, a binary merge tree commencing from the data elements stored at the leaves, which proceeds by merging two by two the nearest subsets until the root of the tree is arrived at.15AC is conceptually easy to understand and execute while producing an informative clustering approach.
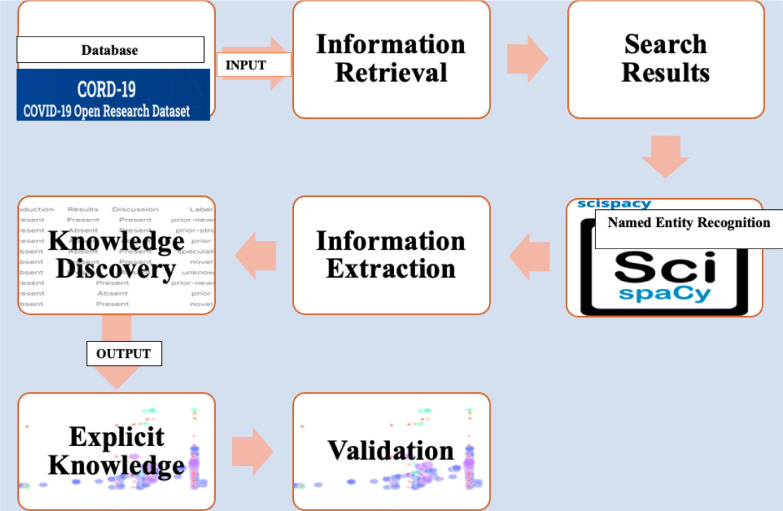
The sequence of our biomedical NLP, which included information retrieval, entity recognition of relevant biomedical entities, medical content information extraction, knowledge discovery of latent relationships, data visualisation (described in the Results section), and validity measures (described in the Results section) is outlined in the fig. 2 .
Fig. 2.
Our biomedical NLP sequence.
Results
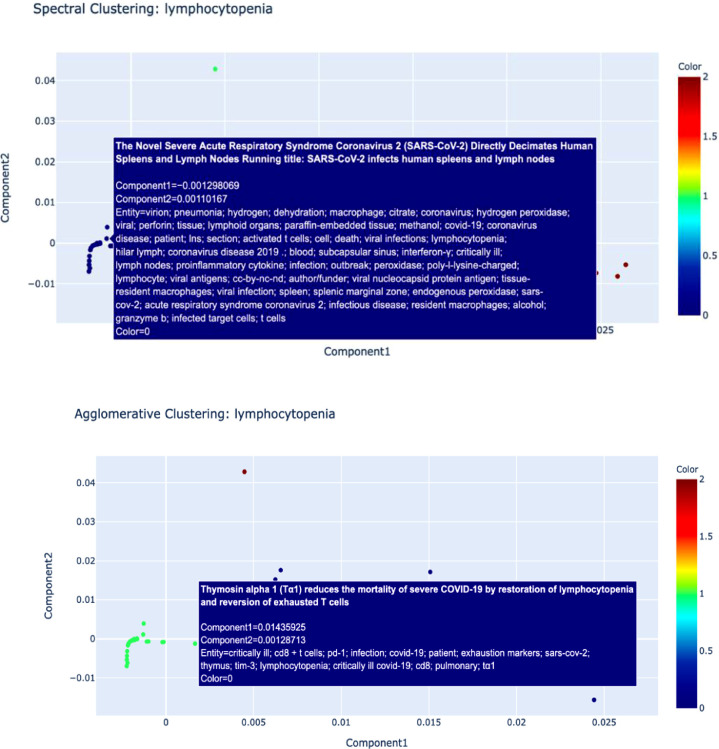
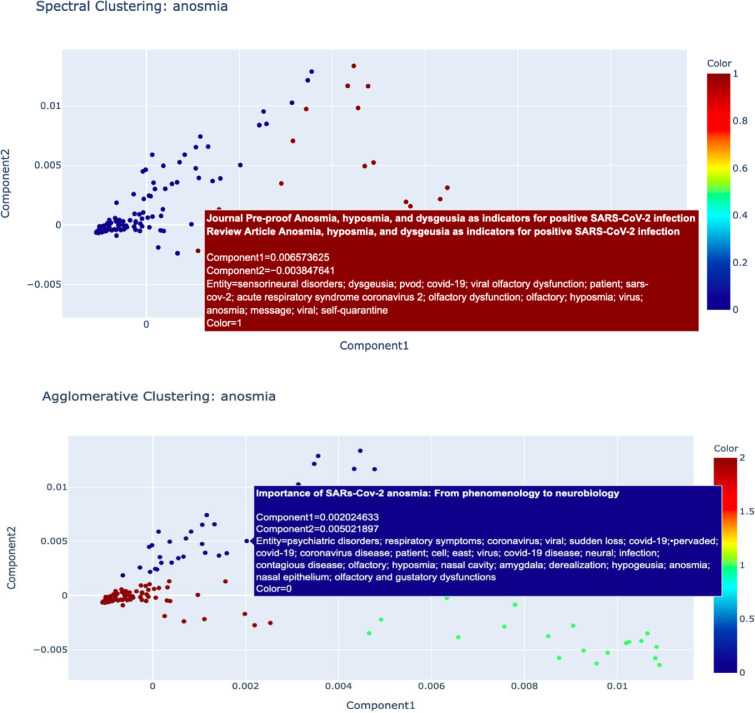
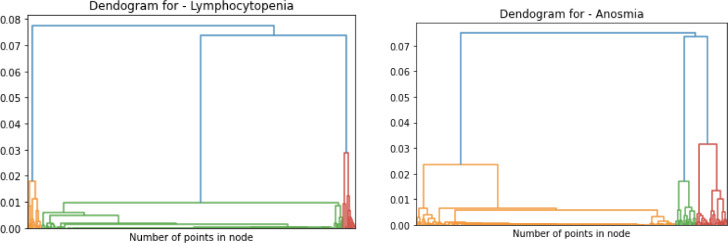
To illustrate the utility of our biomedical NLP, we searched for ‘lymphocytopenia ' and 'anosmia’' terms from the CORD-19 dataset. Both lymphocytopenia and anosmia have been found to be associated with COVID-19 1 , 17 Since the beginning of the pandemic, lymphocyte count has been an important marker of clinical progression of the disease.18 Numerous studies have now shown that lymphocytopenia is associated with poor clinical outcomes for COVID-19 patients. Another prominent sign that has been associated with COVID-19 patients is anosmia, which can present suddenly and sometimes without other symptoms.19 While anosmia has been widely reported as a clinical sign, the pathogenic mechanism of olfactory dysfunction and it clinical characteristics is not clear. Therefore, we considered identifying literature pertaining to these two COVID-19 manifestations would showcase the usefulness of our biomedical data mining model to biomedical and clinical researchers. Outlined below are the representation of the data clusters of the representation including spectral and agglomerative cluster representation of the two terms ‘Lymphocytopenia’ and ‘Anosmia’ (Fig. 3, Fig. 4, Fig. 5 ). In Fig. 3 and 4, the papers relating to the terms are reflected as dots and in Fig. 5, the cluster hierarchy for each search term is presented as a dendrogram (tree diagram).
Fig. 3.
Spectral and Agglomerative Cluster representation for search term ‘Lymphocytopenia'. Each dot represents a paper relating to the term.
Fig. 4.
Spectral and Agglomerative Cluster representation and confusion matrix for search term 'Anosmia’. Each dot represents a paper relating to the term.
Fig. 5.
Dendrogram for agglomerative clustering of search terms ‘Lymphocytopenia’ and 'Anosmia’ respectively.
Validity measures for clustering approaches can be internal and external.20 For this project, we used internal measures as they only rely on information in the data unlike external validation measures, which need information external to the data. As we did not have ground true label of data, we relied just on internal clustering validation. For the quality metrics we used three internal validation measures: Silhouette, Calinski-Harabasz (CH) and Davies-Bouldin (DB) index scores (Table 1 ). Internal validity measures quantify the quality of the clustering relying on properties intrinsic to the dataset.20 , 21 They mathematically measure what a good clustering looks like. They also allow for the comparison of partition between clusters. Silhouette index authenticates the clustering performance based on the pairwise variance of between and within cluster distances.20 By maximising the value of this index, the optimal cluster number can be determined. The CH index, also known as the Variance Ratio, assesses the validity of clusters based on the average between and within cluster sum of squares.22 DB signifies the average similarity between clusters by comparing the distance between clusters with the size of the clusters. In other words it identifies clusters, which are far from each other and compact.23
Table 1.
Quality metrics of dimensionality reduction and cluster coherence.
| Quality Metrics | Spectral Clustering | Agglomerative Clustering | ||
|---|---|---|---|---|
| Search Term | Lymphocytopenia | Anosmia | Lymphocytopenia | Anosmia |
| Silhouette Index Score | 0.890 | 0.833 | 0.929 | 0.894 |
| Calinski-Harabasz Index Score | 196.617 | 256.558 | 258.605 | 849.014 |
| Davies-Bouldin Index Score | 0.808 | 0.898 | 0.514 | 0.539 |
The Silhouette score is bound between -1 and +1 so the higher the score relates to better defined clusters. We notice for both search term ‘lymphocytopenia’ spectral clustering achieves slightly lower performance with Silhouette index than agglomerative clustering indicating that the clusters obtained are not relatively compact. On the other hand, we observed that agglomerative clustering creates tight clusters but well separated both for ‘lymphocytopenia’ and ‘anosmia’. The same pattern recurs for agglomerative clustering with CH scores, which scores dense but well separated clusters higher. Spectral clustering tries to maintain nearest neighbours in reduced dimensions causing the dilution of clusters but still achieving decent CH scores. For agglomerative clustering, DB scores are also better as it forms tight clusters trading off nearest neighbour similarity unlike spectral clustering. This is because DB emphasizes separation which becomes pronounced when inter-cluster distances are very low.
Discussion
Information retrieval, in a text mining context, involves a user submitting a query to a search engine and receiving relevant results aligning with their submitted question in return.24 In a pandemic context, information extraction from medical literature involves the identification of entities such as diseases, as well as the identification of complex relationships between these entities.5 , 6 Query results extracted from the literature may be used to populate databases or data curation.24 From these extractions, knowledge bases can be built that contain the collected statements with references to the literature. Knowledge discovery involves identifying undiscovered or hidden knowledge by applying data-mining algorithms to the collection of facts gathered from the literature. From here, text mining results may be used to suggest new hypotheses which can be used to either validate or disprove existing hypotheses or to help direct future research. 24
The text-mining and clustering visualisation model we have developed assists with the knowledge discovery process by using clustering approaches and uncovering latent relationship between entities aiding researchers and clinicians in their pursuit of appropriate treatment and management of COVID-19 cases. This process is achieved by retrieving articles that mention relevant biomedical terms relating to COVID-19 and categorising them for their relevance to the clinical risk factors. The assumption here is that supplied databases like CORD-19 have relevant information suitable for extraction. A valuable aspect to the model is its information extraction process, which involves both a robust information retrieval engine and biomedical named entity recognition. Knowledge representation was achieved through dimensionality reduction and feature affinity clustering algorithms, which is a novel process not found generally in other biomedical NLP models. Further to this, the garnered findings are presented in visually aesthetic and a readable manner that ensure pertinent insights are conveyed to a broad spectrum of healthcare professionals. While the tool we developed here was customised to identify COVID19 related risk factors, this model can be potentially customised to extract other biomedical terms and assist with knowledge discovery.
Conclusion
To develop the COVID-19 biomedical NLP, we brought together data scientists, software engineers, clinicians, and medical researchers to enable an informed approach and to develop a well-rounded knowledge extraction process. The multi-disciplinary effort emulates the real-world clinical inductive reasoning that utilises a stepped approach to evaluate, extract and prioritise insights to enable evidence-based medicine.
While our biomedical NLP extracts pertinent entities from the biomedical literature and provides a high degree of cluster coherency, it has limitations associated with the biomedical named entity recognition library. This restricts the ability to extract all the necessary biomedical aligned entities. Biomedical concept recognition is an area of active research, and improved methods targeting a broad range of entity and concept types can be substituted. We demonstrated our project to frontline clinicians and the feedback was positive. This is in terms of being able to rapidly access accurate and appropriate information from the COVID infodemic in a visually insightful interface. The COVID-19 pandemic has created a unique situation where there is a need for rapid access to evolving clinical knowledge rapidly from the exponentially increasing publications of variable quality. We strongly believe because of these features the platform may support deeper investigation of the scientific literature related to COVID-19.
Declaration of competing interest
None.
Acknowledgments
Data availability
The CORD-19 dataset used to develop our biomedical NLP tool can be accessed here: https://www.semanticscholar.org/cord19/download.
Code availability
The code used for this study is available at: https://github.com/ravibhaskar/CORD-19-S4PCKR.
Author contributions
All authors contributed to the development and validation of the model described in the paper, and the writing of this paper.
References
- 1.Guan W.J., et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chowell G., Mizumoto K. The COVID-19 pandemic in the USA: what might we expect? Lancet. 2020;395:1093–1094. doi: 10.1016/S0140-6736(20)30743-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA - J. Am. Med. Assoc. 2019 doi: 10.1001/jama.2020.6019. 2020. [DOI] [PubMed] [Google Scholar]
- 4.NIH. iSearch COVID-19 Portfolio. https://icite.od.nih.gov/covid19/search/(2020).
- 5.Brainard J. Scientists are drowning in COVID-19 papers. Can new tools keep them afloat? Science. 2020 doi: 10.1126/science.abc7839. https://www.sciencemag.org/news/2020/05/scientists-are-drowning-covid-19-papers-can-new-tools-keep-them-afloat [DOI] [Google Scholar]
- 6.Zhu F., et al. Biomedical text mining and its applications in cancer research. J. Biomed. Inform. 2013;46:200–211. doi: 10.1016/j.jbi.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Allen Institute for AI COVID-19 Open Research Dataset. Semantic Scholar. 2020 https://www.semanticscholar.org/cord19 [Google Scholar]
- 8.Apache . 2020. Solr is the popular, blazing-fast, open source enterprise search platform built on Apache Lucene.https://lucene.apache.org/solr/ [Google Scholar]
- 9.Neumann, M., King, D., Beltagy, I. & Ammar, W. ScispaCy: fast and robust models for biomedical natural language processing. 319–327 (2019) doi: 10.18653/v1/w19-5034. [DOI]
- 10.Hong L., et al. A novel machine learning framework for automated biomedical relation extraction from large-scale literature repositories. Nat. Mach. Intell. 2020;2:347–355. [Google Scholar]
- 11.Nie F., Xu D., Tsang I.W., Zhang C. Proceedings of the 21st international joint conference on Artificial Intelligence. 2009. Spectral Embedded Clustering; pp. 1181–1186. [Google Scholar]
- 12.Kim J.H., Kohane I.S., Ohno-Machado L. Visualization and evaluation of clusters for exploratory analysis of gene expression data. J. Biomed. Inform. 2002;35:25–36. doi: 10.1016/s1532-0464(02)00001-1. [DOI] [PubMed] [Google Scholar]
- 13.Ng, A. Y., Jordan, M. I. & Weiss, Y. On spectral clustering: analysis and an algorithm. in NIPS 01: Proceedings of the 14th International Conference on Neural Information Processing Systems:Natural and Synthetic 14 (2001). doi:10.1.1.19.8100.
- 14.Von Luxburg U. A tutorial on spectral clustering. Stat. Comput. 2007;17:395–416. [Google Scholar]
- 15.Nielsen F. Hierarchical clustering. in Introduction to HPC with MPI for Data Science. Springer International Publishing. 2016;304 doi: 10.1007/978-3-319-21903-5. [DOI] [Google Scholar]
- 16.Zhang W., Wang X., Zhao D., Tang X. vol. 7572. 2012. Graph degree linkage: Agglomerative clustering on a directed graph; pp. 428–441. (Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics)). LNCS. [Google Scholar]
- 17.Liao D., et al. Haematological characteristics and risk factors in the classification and prognosis evaluation of COVID-19: a retrospective cohort study. Lancet. Haematol. 2020;3026:1–8. doi: 10.1016/S2352-3026(20)30217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang I., Pranata R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): systematic review and meta-analysis. J. Intensive Care. 2020;8:1–10. doi: 10.1186/s40560-020-00453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng X., Deng Y., Dai Z., Meng Z. COVID-19 and anosmia: a review based on up-to-date knowledge. Am J Otolaryngol. 2020;41:19–21. doi: 10.1016/j.amjoto.2020.102581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, Y., Li, Z., Xiong, H., Gao, X. & Wu, J. Understanding of internal clustering validation measures. Proc. - IEEE Int. Conf. Data Mining, ICDM 911–916 (2010) doi: 10.1109/ICDM.2010.35. [DOI]
- 21.Van Craenendonck T., Blockeel H. Using internal validity measures to compare clustering algorithms. Icml. 2015:1–8. [Google Scholar]
- 22.Wei H. How to measure clustering performances when there are no ground truth? Medium. 2020 https://medium.com/@haataa/how-to-measure-clustering-performances-when-there-are-no-ground-truth-db027e9a871c [Google Scholar]
- 23.Saitta S., Raphael B., Smith I.F.C. A comprehensive validity index for clustering. Intell. Data Anal. 2008;12:529–548. [Google Scholar]
- 24.Fleuren W.W.M., Alkema W. Application of text mining in the biomedical domain. Methods. 2015;74:97–106. doi: 10.1016/j.ymeth.2015.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The CORD-19 dataset used to develop our biomedical NLP tool can be accessed here: https://www.semanticscholar.org/cord19/download.