Figure 1.
Global transcript profiling of mouse tissues over 24 h reveals robust diurnal rhythms of transcriptomes under time-restricted feeding
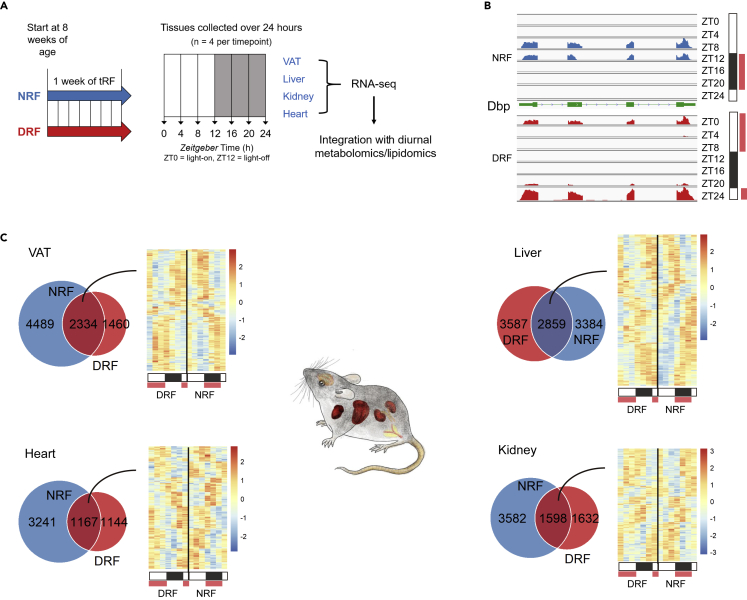
(A) The workflow of multi-omics analysis of diurnal rhythms in peripheral tissues from female mice subjected to inverted feeding for 1 week. All transcript profiles have at least 40 million reads per sample. DRF, daytime-restricted feeding; NRF, nighttime-restricted feeding; VAT, visceral adipose tissue. See transparent methods.
(B) Read counts from the clock output gene Dbp locus exhibit diurnal rhythms in liver and are phase inverted under DRF. ZT, zeitgeber time. See also Figures S1A and S1B.
(C) General features of the global transcript profiling of peripheral tissues in 9-week-old time-restricted-fed C57BL/6J female mice. Venn diagrams illustrate the overlap of oscillating genes between DRF and NRF in each tissue. Heatmaps show 24-h expression profiles of genes that oscillated under both DRF and NRF conditions.