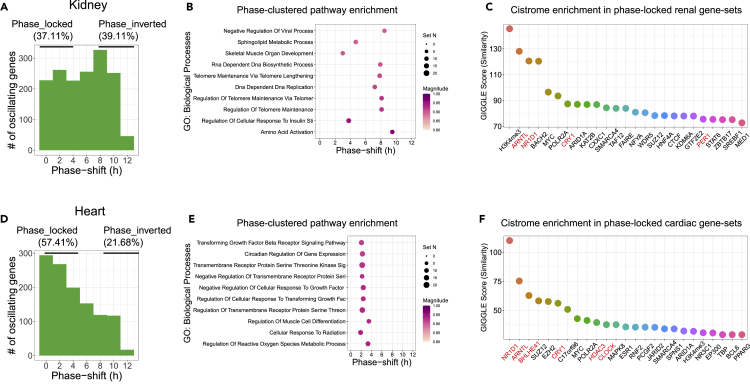
Figure 5.
Diurnal transcriptomes in kidney and heart show slow kinetics/resistance in phase entrainment to feeding
(A) Phase-shift histogram shows that 39.11% of the renal dual-oscillating genes were phase inverted by inverted feeding. Phase-locked, phase shift of 0–4 h; phase-inverted, phase shift of 8–12 h. Considering the cyclic nature, phase shift is converted to absolute value to allow a window of 0–12 h, e.g. a phase advance of 14 h equals to a phase delay of 10 h (24–14 = 10), and is expressed as a phase shift of 10 h. See also Figure S5A.
(B) Top 10 phase-clustered pathways that are phase shifted by inverted feeding in kidney. Magnitude is a measure of the temporal cohesiveness of the phase set (q < 0.05).
(C) Cistrome enrichment analysis of phase-locked genes in kidney based on curated cistromes of transcription factors and chromatin regulators from CistromeDB. Circadian clock proteins are marked in marked. See also Figure S5B.
(D) Phase-shift histogram shows that 21.68% of the cardiac dual-oscillating genes were phase inverted by inverted feeding. See also Figure S5C.
(E) Top 10 phase-clustered pathways that are phase shifted by inverted feeding in heart.
(F) Cistrome enrichment analysis of phase-locked genes in heart based on curated cistromes of transcription factors and chromatin regulators from CistromeDB. Circadian clock proteins are marked in red. See also Figure S5D.