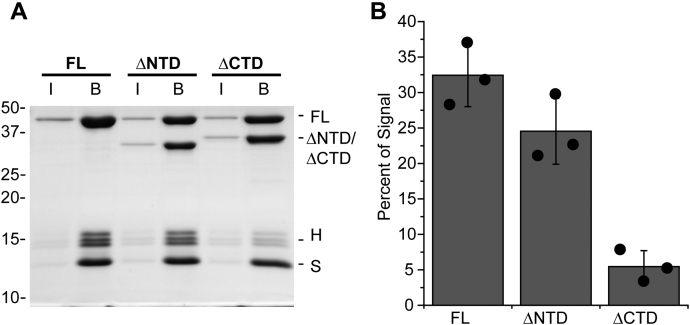
Figure 5.
Affinity of PFV IN truncation mutants for 601 nucleosomes in the presence of physiologically relevant monovalent salt concentration.A, FL, PFV IN(ΔNTD), and PFV IN(ΔCTD) intasomes were assembled with vDNA labeled with biotin. The intasomes were added to 601 nucleosomes and streptavidin-conjugated beads in the presence of 110 mM NaCl. The beads were extensively washed, analyzed by PAGE, and stained with Coomassie brilliant blue. Lane I, 5% of the total protein. Lane B, proteins associated with beads. Histones H3, H2B, and H2A (H). Streptavidin (S) and histone H4 have the same mobility. B, the total Coomassie signal in each lane was calculated, excluding the band of streptavidin and H4. The percentage of the total H3, H2B, and H2A signal in each lane B was calculated. Black circles indicate values from each experiment. Error bars indicate the standard deviation between three experiments with at least two independent PFV intasome and nucleosome preparations.