FIGURE 1.
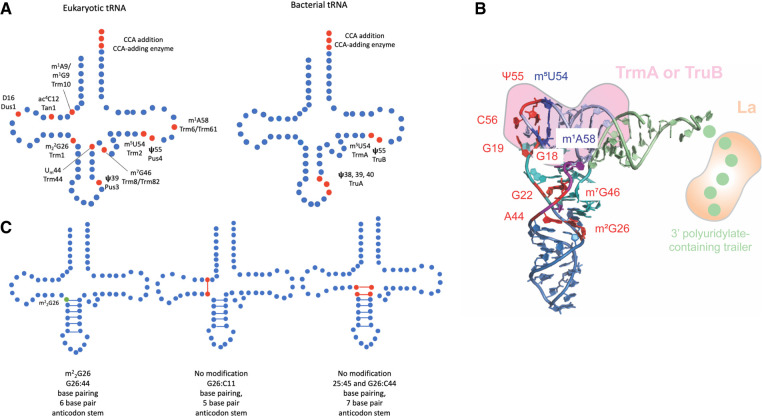
(A) Schematic of eukaryotic and bacterial tRNA modifications discussed and the respective enzymes responsible for each modification. Modified nucleotides are indicated in red. (B) Three-dimensional tRNA structure highlighting tertiary base pairs and interaction sites of tRNA chaperones. The different tRNA arms are color-coded with the acceptor arm in light green, the D arm in cyan, the acceptor arm in blue, the variable arm in purple, and the T arm in light blue (structure 4TRA from Westhof et al. 1988), visualized in PyMOL (The PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC). Critical tertiary interactions involving modified nucleotides connecting different tRNA arms are highlighted in red: m2G26—A44 (hinge region), G22—m7G46, G18—Ψ55, and G19—C56. The tertiary interaction of m5U54 and m1A58 in the T-loop is depicted in blue. La (orange) binds to the 3′ polyuridylate-containing trailer. TrmA and TruB (pink) both interact with the T arm causing the disruption of tertiary base pairs to the D arm. (C) The influence of m22G26 modification on base-pairing and anticodon length (Steinberg and Cedergren 1995). m22G26 modification is indicated in green and noncanonical base pairs resulting from unmodified G26 are indicated in red.