Figure 3.
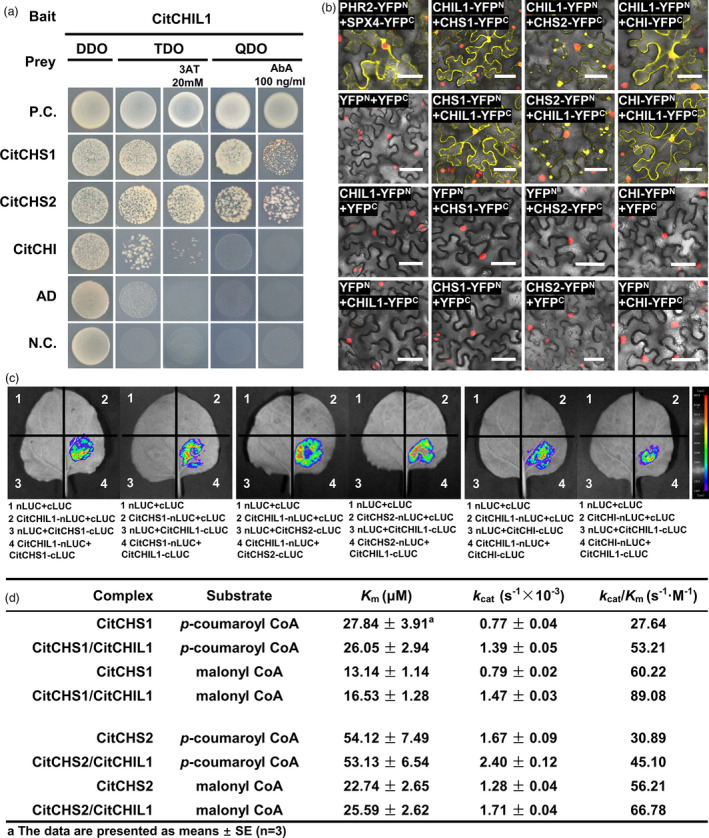
Physical interaction between CitCHIL1 and other EBG‐encoded proteins. (a) Yeast two‐hybrid analyses of the protein–protein interaction between CitCHIL1 and CitCHS1, CitCHIL1 and CitCHS2, CitCHIL1 and CitCHI. The TDO medium (SD‐Trp‐Leu‐His medium) and QDO medium (SD‐Trp‐Leu‐His‐Ade medium) were used as selective media. 3AT, 3‐amino‐1,2,4‐triazole; AbA, aureobasidin A; P.C., positive control, pGBKT7‐53 and pGADT7‐T constructs; AD, pGBKT7‐CitCHIL1 and pGADT7 constructs; N.C., negative control, pGBKT7 and pGADT7 constructs. (b) Bimolecular fluorescence complementation (BiFC) analyses of the protein–protein interactions between CitCHIL1 and CitCHS1, CitCHIL1 and CitCHS2, CitCHIL1 and CitCHI in transgenic Nicotiana benthamiana leaves (expressed together with nucleus‐located mCherry). The pairs of fusion proteins tested were CitCHIL1‐YFPN + CitCHI‐YFPC, CitCHI‐YFPN + CitCHIL1‐YFPC, CitCHIL1‐YFPN + CitCHS1‐YFPC, CitCHS1‐YFPN + CitCHIL1‐YFPC, CitCHIL1‐YFPN + CitCHS2‐YFPC, and CitCHS2‐YFPN + CitCHIL1‐YFPC. The pair PHR2‐YFPN + SPX4‐YFPCwas a positive control. The other combinations were negative controls. The yellow fluorescence visualized the interaction in vivo. Bars = 50 μm. (c) Firefly luciferase complementation imaging (LCI) analyses of the direct interaction between CitCHIL1 and CitCHS1, CitCHIL1 and CitCHS2, CitCHIL1 and CitCHI in Nicotiana benthamiana leaves. The luciferase images visualized the interaction in vivo. (d) Kinetic parameters of CitCHSs or CitCHIL1/CitCHS1 and CitCHIL1/CitCHS2 complexes with p‐coumaroyl‐CoA and malonyl‐CoA as substrates. For kinetic determination, malonyl‐CoA at a fixed concentration of 200 μm and p‐coumaroyl‐CoA at a series of concentrations (3–180 μm), orp‐coumaroyl‐CoA with a fixed concentration of 60 μm and malonyl‐CoA at a series of concentration (2–180 μm), were added to the standard CHS reactions. The K m and V max values were calculated by nonlinear regression curves fitted to the Michaelis–Menten equation using GraphPad Prism 8.0.
