Figure 3.
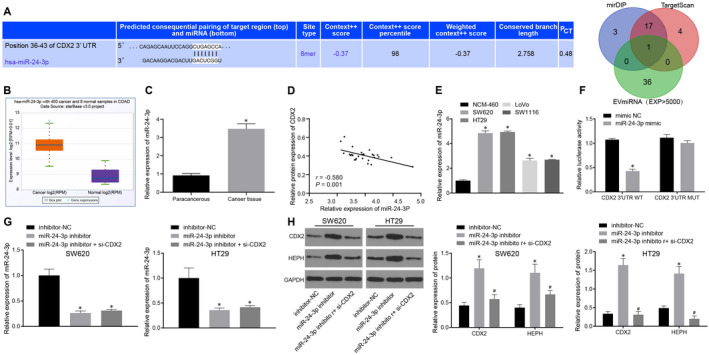
miR‐24‐3p is overexpressed in CC tissues and cells and CDX2 is a target gene of miR‐24‐3p. A, Prediction of upstream miRNA of CDX2. The 3 circles in the FIGURE represent the prediction results of the mirDIP and TargetScan database, and the miRNAs in the fibroblast microbubbles obtained on the EVmiRNA database (lower FIGURE). The binding site between miR‐24‐3p and CDX2 predicted on the TargetScan database (upper FIGURE). The middle part represents the intersection of the three sets of data. B, Expression of miR‐24‐3p in CC on TCGA. The red box plot represents the tumour samples, and the purple box plot represents the normal samples. C, The expression of miR‐24‐3p in CC tissues and adjacent tissues measured by RT‐qPCR normalized to U6. D, Spearman's correlation test of correlation between CDX2 mRNA expression and miR‐24‐3p expression in clinical samples of CC tissues. E, The expression of miR‐24‐3p in NCM‐460 cells and CC cells (SW620, HT29, LoVo and SW1116) examined by RT‐qPCR normalized to U6. F, The binding relationship between miR‐24‐3p and CDX2 assessed by dual‐luciferase reporter gene assay. G, The expression of miR‐24‐3p in cells after inhibition of miR‐24‐3p and CDX2 detected by RT‐qPCR normalized to U6. H, The protein expression of CDX2 and HEPH in cells after inhibition of miR‐24‐3p and CDX2 measured by Western blot analysis normalized to GAPDH. * P < .05 vs adjacent tissues, NCM‐460 cells or inhibitor‐NC‐transfected cells. # P < .05 vs miR‐24‐3p inhibitor–transfected cells. Measurement data were expressed as mean ± standard deviation. Data of clinical samples were compared using paired t test, and data between cells were compared by unpaired t test. Data between two groups were compared by paired t test, and comparisons among multiple groups were conducted by one‐way analysis of variance (ANOVA), followed by Bonferroni's post hoc test
