Fig. 4. Functional analysis of full-length ADAMTS13 CUB1-2 mutants.
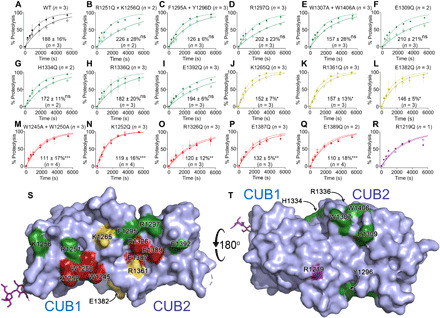
(A) Recombinant full-length ADAMTS13 was analyzed functionally using VWF96 as a substrate. Proteolysis of VWF96 by 0.45 nM ADAMTS13 in the absence (solid line) and presence (dotted line) of the activating anti-CUB domain monoclonal antibody 17G2 that disrupts the Spacer-CUB interaction was quantified over time by ELISA. Progress curves are shown as mean ± SEM (n = 3). In the presence of 17G2, ADAMTS13 catalytic efficiency is enhanced by 188 ± 13% (mean ± SD; n = 4). (B to R) Parallel analysis of recombinant ADAMTS13 CUB domain mutants. The number of replicates for each dataset presented in the graph is given. The % enhancement effect of 17G2 ± SD is shown for each mutant with the number of replicates (including reactions performed at different enzyme concentrations that are not represented graphically). (B) to (I) are mutants shown in green that are activated similar to wild-type (WT) ADAMTS13. The % activation is not significantly different (ns) to WT ADAMTS13. (J) to (L) are mutants shown in yellow that exhibit partial, but significantly reduced activation in response to 17G2 (*P ≤ 0.05). (M) to (Q) are mutants shown in red that exhibit highly significantly reduced activation in response to 17G2 (**P ≤ 0.01; ***P ≤ 0.001). (R) ADAMTS13 R1219Q mutant was not activated by 17G2, but this was subsequently found to be due to the loss of binding of the 17G2 antibody. All other mutants bound 17G2 normally. (S and T) ADAMTS13 CUB1-2 domain surface amino acids and their influence upon the Spacer-CUB interaction. Amino acids depicted in fig. S4 are color-coded according to their influence on the Spacer-CUB interaction. Amino acids shown in green do not influence the Spacer-CUB interaction, those in yellow have a moderate or indirect effect, whereas those residues highlighted in red that form two adjacent patches in CUB1 and CUB2 represent amino acids that have a major influence upon Spacer-CUB binding.
