Fig. 5. Proposed interaction between the ADAMTS13 CUB1-2 domains and the ADAMTS13 Spacer domain.
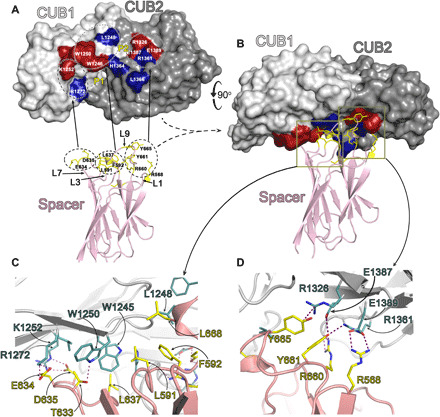
(A) Space-fill representation of the ADAMTS13 CUB1-2 domains with residues identified from functional analyses to influence the Spacer-CUB interaction labeled in red. Those labeled in blue are amino acids predicted from the refined ClusPro docking simulation to contribute to Spacer domain binding. The positions of the P1 and P2 pockets are shown. Below is the ADAMTS13 Spacer domain that interacts with the CUB domains shown in cartoon representation. In stick representation (yellow) are the reciprocal residues that likely interact directly with the CUB1-2 domain patches. The Spacer domain loop (L) numbering for the highlighted residues is given. (B) Favored docking model of the CUB1-2 domains interacting with the Spacer domain. Note that the CUB1-2 domains have rolled forward by ~90° when compared to (A). Boxed areas highlight the two areas enlarged in (C) and (D), corresponding to the CUB1 and CUB2 interaction sites, respectively. (C) CUB1 interaction site with the Spacer domain. The R-groups of amino acids involved in either hydrophobic or ionic interactions are shown in stick representation and labeled (CUB residues in teal; Spacer residues in yellow). (D) CUB2 interaction site with the Spacer domain. The R-groups of amino acids involved in ionic interactions are shown in stick representation and labeled (CUB residues in teal; Spacer residues in yellow).
