Table 1.
Docking table with bonding characterization and binding energies in kcal/mol with 14 active compounds involved in five therapeutic targets.
| No. | Name | Structure | Glide G-score | MM-GBSA dG bind (kcal/mol) | Bonding interaction | Bond type | Binding protein |
|---|---|---|---|---|---|---|---|
| 4 | Polydatin |

|
−8.846 | −44.64 | ASP A 158, GLU A 62, GLU B 360, ASN A 145, SER A 144 | H-acc | MAPK1 (PDB ID: 5ax3 native ligand G-score: −8.713) |
| TYR A 27, GLN A 45 | H-don | ||||||
| LYS A 45 | Pi-cation | ||||||
| 5 | Resveratrol |
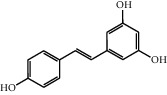
|
−7.539 | −37.18 | GLN B 192, SER B 530 | H-acc | PTGS2 (PDB ID: 5kir native ligand G-score: −9.800) |
| HIE B 90 | H-don | ||||||
| TRP B 387 | Pi-pi stacking | ||||||
| 10 | Rutin |

|
−7.320 | −53.30 | SER A 68 | H-acc | LCN2 (PDB ID: 1x89 native ligand G-score: −7.190) |
| ARG A 81 | H-don | ||||||
| TYR A 106 | Pi-pi stacking | ||||||
| LYS A 125 | Pi-cation | ||||||
| 11 | Kaempferol |

|
−7.734 | −47.73 | ALA B 191, GLY B 233 | H-acc | MMP9 (PDB ID: 5ue4 native ligand G-score: −8.661) |
| HIS B 230 | H-don | ||||||
| PHE B 110 | Pi-pi stacking | ||||||
| 12 | Dihydrokaempferol |

|
−8.167 | −32.82 | GLU A 62, ASP A 158, GLU A 24, MET A 99 | H-acc | MAPK1 |
| GLN A 96 | H-don | ||||||
| LYS A 45 | Pi-cation | ||||||
| 17 | Quercetin |
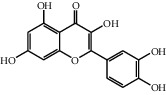
|
−8.367 | −38.06 | ASP A 158, GLU B 360, GLU A 62 | H-acc | MAPK1 |
| GLN A 96 | H-don | ||||||
| LYS A 45 | Pi-cation | ||||||
| 18 | Isorhamnetin |
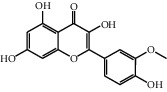
|
−9.757 | −51.06 | SER B 530, GLN B 192 | H-acc | PTGS2 |
| PHE B 518, TYR B 385 | H-don | ||||||
| 21 | Piceatannol |

|
−8.673 | −40.61 | GLU B 360, GLU A 62, ASP A 158, | H-acc | MAPK1 |
| GLN A 96 | H-don | ||||||
| LYS A 45 | Pi-cation | ||||||
| 30 | Oxyresveratrol |
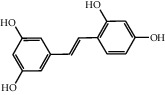
|
−9.241 | −56.71 | ALA B 191, HIS B 230, GLY B 233 | H-acc | MMP9 |
| PHE B 110, TYR B 179 | Pi-pi stacking | ||||||
| 44 | Moracin M |

|
−9.326 | −37.01 | GLU A 62, GLU B 360, MET A 99 | H-acc | MAPK1 |
| GLN A 96 | H-don | ||||||
| 48 | Dihydroquercetin |

|
−7.020 | −41.83 | TYR D 151 | H-acc | TNF (PDB ID: 2az5 native ligand G-score: −7.879) |
| TYR D 59 | Pi-pi stacking | ||||||
| 58 | Kaempferide |

|
−7.962 | −47.23 | GLY B 233, ALA B 191 | H-acc | MMP9 |
| HIS B 230 | H-don | ||||||
| PHE B 110 | Pi-pi stacking | ||||||
| 60 | Gramine |
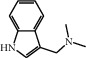
|
−7.452 | −30.51 | GLY A 121 | H-acc | TNF |
| 62 | Dihydrokaempferide |
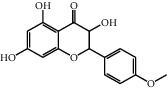
|
−7.638 | −38.65 | GLN B 192 | H-acc | PTGS2 |
| TYR B 385 | H-don |
MM-GBSA dG bind: the binding energy of the receptor and ligand as calculated by the prime energy, a molecular mechanics + implicit solvent energy. Function (kcal/mol) = prime energy (optimized complex) − prime energy (optimized free ligand) − prime energy (optimized free receptor).
