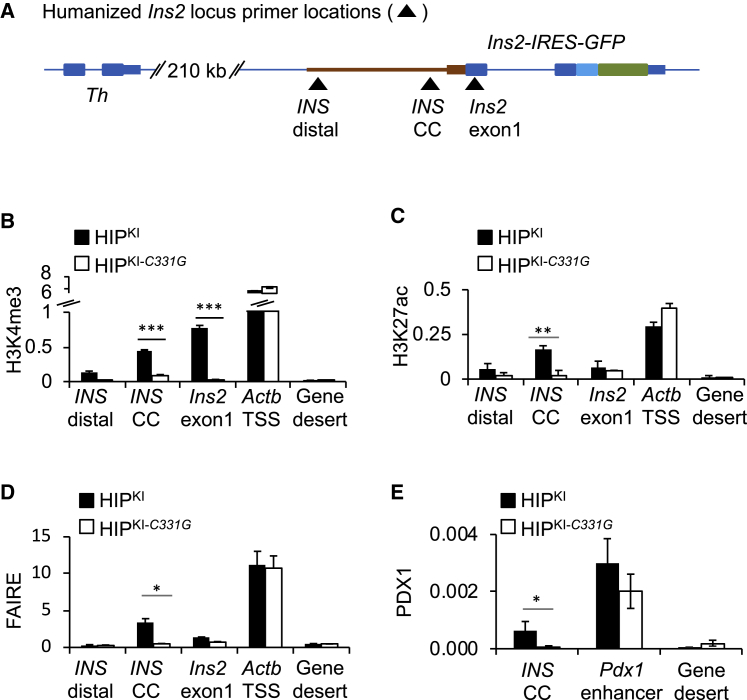
Figure 4.
HIPKI recapitulates and HIPKI-C331G abrogates formation of active chromatin at the INS locus
(A) Schematic showing approximate primer locations for ChIP assays. INS CC primers encompass the CC element.
(B–D) ChIP quantification of H3K4me3, H3K27ac, and FAIRE accessibility at the humanized INS locus and control regions in HIPKI or HIPKI-C331G adult mouse islets. The Actb promoter was used as a positive control, and a gene desert lacking active histone modifications in islets was used as a negative control. ChIP and FAIRE DNA were expressed as a percentage of the total input DNA. n = 3 independent experiments.
(E) ChIP for the transcription factor PDX1 in HIPKI or HIPKI-C331G adult mouse islets at a known PDX1-bound enhancer near Pdx1 as a positive control. Data are expressed as a percentage of input DNA. n = 3 independent experiments. Error bars indicate SEM; asterisks indicate Student’s t test p values (∗p < 0.05; ∗∗p < 0.001; ∗∗∗p < 0.0001).