Abstract
BACKGROUND:
The placenta is formed by the coordinated expansion and differentiation of trophoblast stem (TS) cells along a multi-lineage pathway. Dynamic regulation of histone 3 lysine 9 (H3K9) methylation is pivotal to cell differentiation for many cell lineages, but little is known about its involvement in trophoblast cell development.
METHODS:
Expression of H3K9 methyltransferases was surveyed in rat TS cells maintained in the stem state and following differentiation. The role of suppressor of variegation 3–9 homolog 2 (SUV39H2) in the regulation of trophoblast cell lineage development was investigated using a loss-of-function approach in rat TS cells and ex vivo cultured rat blastocysts.
RESULTS:
Among the twelve-known H3K9 methyltransferases, only SUV39H2 exhibited robust differential expression in stem versus differentiated TS cells. SUV39H2 transcript and protein expression were high in the stem state and declined as TS cells differentiated. Disruption of SUV39H2 expression in TS cells led to an arrest in TS cell proliferation and activation of trophoblast cell differentiation. SUV39H2 regulated H3K9 methylation status at loci exhibiting differentiation-dependent gene expression. Analyses of SUV39H2 on ex vivo rat blastocyst development supported its role in regulating TS cell expansion and differentiation. We further identified SUV39H2 as a downstream target of caudal type homeobox 2, a master regulator of trophoblast lineage development.
CONCLUSIONS:
Our findings indicate that SUV39H2 contributes to the maintenance of TS cells and restrains trophoblast cell differentiation.
GENERAL SIGNIFICANCE:
SUV39H2 serves as a contributor to the epigenetic regulation of hemochorial placental development.
Keywords: SUV39H2, histone H3K9 methylation, trophoblast stem cell differentiation, hemochorial placentation
1. Introduction
Development of the trophoblast cell lineage is a key event required for growth and survival of the embryo in the female reproductive tract (1–3). This developmental process requires a sequential progression: i) conversion of pluripotent cells of the early embryo into a multipotent trophoblast cell lineage, ii) expansion of trophoblast stem (TS) cells, and iii) differentiation of trophoblast cell lineages, including cell types with a specialized capacity to modify the uterine environment and a barrier function directly controlling nutrient flow to the fetus (4). Disruptions in trophoblast cell lineage development can result in embryo implantation failure and pregnancy-related disorders (4). Understanding mechanisms controlling trophoblast cell development has considerable relevance in successful pregnancy outcomes. Transcription factors, histone modifications, and chromatin organizers have been implicated as regulators of this fundamental process (3, 5, 6). It is expected that transcription factors target sets of genes pivotal to trophoblast cell lineage development through specific DNA nucleotide recognition motifs and recruit various co-regulators, including enzymes capable of modifying histones and chromatin structure, through the assistance of chromatin organizers. Alternative delivery systems may also exist, including long noncoding RNAs, which have been implicated in trafficking of chromatin modifying complexes to specific loci within the genome (7). Histone post-translational modifications can also contribute to the establishment euchromatic and heterochromatic regions within the genome during embryonic development (8, 9).
Histone 3 lysine 9 (H3K9) can be monomethylated (H3K9me1), dimethylated (H3K9me2), or trimethylated (H3K9me3) (10). H3K9 methyl transferases catalyze the addition of methyl groups to H3K9, while their removal is catalyzed by H3K9 demethylases. In general, histone H3K9 methylation is associated with three events that direct the organization and function of the genome: i) formation of heterochromatin, ii) DNA methylation, and iii) gene repression (10). Histone H3K9 methylation and/or H3K9 methylation enzymatic machinery are known regulators of cell differentiation, including embryonic stem cells, extraembryonic endoderm, neural crest cells, neuronal cells, mammary epithelial cells, and myoblasts (11–20).
A few studies have implicated H3K9 methylation status in the regulation of trophoblast cell development. During blastocyst cell fate determination SET domain bifurcated histone lysine methyltransferase (SETDB1) contributes to the restriction of trophectoderm development (21–23). In TS cells, the H3K9 methyl transferase, suppressor of variegation 3–9 homolog 1 (SUV39H1), is involved in establishing histone H3K9me3 marks at the promoters of a set of pluripotency genes and is correlated with repression of their expression (24). H3K9 methyl transferases (SETDB1, SUV39H1, SUV39H2) have also been implicated in early porcine embryonic growth and blastocyst formation (25). Rossant and co-workers showed that promoter regions of a select group of genes differentially activated in the TS cell state versus the differentiated state underwent changes in their H3K9me3 status (26). Histone H3K9me3 was associated with transcriptional repression in TS cells. Furthermore, the histone H3K9 demethylase, lysine demethylase 3A (KDM3A), mediates hypoxia-driven trophoblast cell differentiation (27).
In this study, we explore the involvement of SUV39H2 in the regulation of TS cell fate. Targets for SUV39H2 action are identified in TS cells. In vitro and ex vivo research strategies were performed utilizing TS cells and lentiviral trophoblast-specific gene manipulation, respectively. Our experimental findings demonstrate a role for SUV39H2 in the maintenance of TS cells and inhibition of trophoblast cell differentiation.
2. Materials and methods
2.1. Animals and tissue collection
Holtzman Sprague Dawley rats were purchased from Envigo (Indianapolis, IN). Animals were housed in an environmentally controlled facility with lights on from 0600–2000 h and allowed free access to food and water. Timed mating was done by cohabiting virgin female rats 8–10 weeks of age and adult males (>3 months of age) of the same strain. The presence of sperm in the vaginal lavage was considered gestation day (gd) 0.5.
Rat placental tissues were collected, and placentation sites dissected as previously described (28). Tissues for histological analysis were frozen in dry-ice cooled heptane and stored at −80°C. Tissue samples for protein or RNA extraction were frozen in liquid nitrogen and stored at −80°C until processed. The University of Kansas Medical Center (KUMC) Animal Care and Use Committee approved protocols for the care and use of animals.
Paraffin-embedded human placental tissue was obtained from the Research Centre for Women’s and Children’s Health Biobank (Mount Sinai Hospital, Toronto). Tissue collections were performed with consent and were approved by the University of Toronto and the KUMC human research ethics review committees.
2.2. Rat TS cell cultures
Rat TS cells (29) were cultured in TS Cell Complete Medium [RPMI 1640 (Cellgro, Herndon, VA), 20% fetal bovine serum (FBS; Atlanta Biologicals, Norcross, GA), 100 μM 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO), 1 mM sodium pyruvate (Cellgro, Herndon, VA), 50 μM penicillin and 50 U/ml streptomycin (Cellgro)] supplemented with 70% rat embryonic fibroblast conditioned medium (REF-CM), fibroblast growth factor 4 (FGF4, 25 ng/ml; Sigma-Aldrich) and heparin (1 μg/ml; Sigma-Aldrich). TS cells are induced to differentiate by removing FGF4, heparin, and REF-CM from the TS Cell Complete Medium (29).
2.3. Reverse transcription-quantitative PCR (RT-qPCR)
Extraction of total RNA from all samples was carried out using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. First strand cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen) and diluted five times with water and subjected to RT-qPCR. Primer sequences can be found in Table 1. RT-qPCR was performed on an ABI Prism 7500 real time PCR system (Applied Biosystems, Foster City, CA). Each reaction was set up containing SYBR GREEN PCR Master Mix (Applied Biosystems) and primers (250 nM each) in a 10 μl reaction. Cycling parameters included an initial holding step (95 °C for 10 min) and 40 cycles of a two-step PCR (92 °C for 15 sec, then 60 °C for 1 min), followed by a dissociation step (95 °C for 15 sec, 60 °C for 15 sec, and then 95 °C for 15 sec). The ΔΔCt method was used for relative quantification of the amount of mRNA. Each sample was normalized to 18S RNA.
Table 1.
Primers used for RT-qPCR.
| Transcript | Forward | Reverse |
|---|---|---|
| Suv39h2 | ATGAGTTCACAGTGGATG | GCCGAGTATCAAGGTTAT |
| Suv39h1 | GTGACCGTTACCCTTTCGGT | AGCACACACTGCAACCTAGA |
| Setdb1 | TGCCCACAGGGGTTTATGAG | AATGTCATCCAAGCAGCGGA |
| Setdb2 | GGGCCGTTTCCTTAATTTGCT | AATGTCATCCAAGCAGCGGA |
| Ehmt1 | CTGATCGTGCTGCAAAGATGG | GAACTCAGGTCAGACTCATCTCC |
| Ehmt2 | CACCCTTACCGACAACGAGG | CCCGTGGTAGTTGACAGCAT |
| Prl3b1 | ACCATGCTTCTCTGGGACACT | AGGCTTCCAGTGGACATTCGGTAA |
| Chd9 | TGGGGAGATGTTGTTAAGCAGT | CTTTTAGGCCTTCTCCCCCG |
| Foxo4 | TCATTTCCTGTGTCAGCGCC | CCTTCGACTTTCCGCCTCTC |
| Mmp12 | GCTGGTTCGGTTGTTAGG | GTAGTTACACCCTGAGCATAC |
| Cyp11a1 | ACCTATTCCGCTTTGCCTTT | CATGTTGAGCATGGGAACAC |
| Prl2a1 | GCTTCCAAAACCCAGCAGTA | AAGGATGGCAGGTTGTTCAC |
| Prl5a1 | TCCACACCAGACATTCCAGA | TTTCCAGGAAGCCAACATTC |
| Adm | CCCTGATGTTATTGGGTTCG | AGTTCCCTCTTCCCACGACT |
| Cited2 | GAAGGACTGGAAATGGCAGA | GCGCCGTAGTGTATGTGCT |
| Dnmt3l | CCCCAGATCTGAAAGAGGAAGAC | CGTACCTGATGACCTCTCTGC |
| Elf5 | CAAGACTGTCACAGCCGAACAA | TTCTTCCTTTGTCCCCACATC |
| Phlda2 | TGGCGCTGATCGACTACCA | ATACCTGAAGCGGCGAAACTC |
| Id2 | GGACATCAGCATCCTGTCCT | AAAAAGGAAAAAGTCCCCAAA |
| Cdx2 | CAGGAGGAAAGCTGAGTTGG | TGCTGCTGTTGCAACTTCTT |
| Bmp4 | GAAGAACATCTGGAGAACATC | GGGCTTCATAACCTCATAAAT |
| Prl3d1 | TCGCGCCTCTGGTATGCAAC | TGGACACAATGGCAGTTGGTTTGG |
| 18S | GCAAGAGCAGAAGGGTAAAGAAGG | GGCCTCACTAAACCATCCAA |
2.4. Western blot analysis
Rat TS cell lysates were prepared in radioimmunoprecipitation assay buffer (10 mM Tris-HCl, pH 7.2, 1% Triton X-100 or 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 5 mM EDTA, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin). Protein concentrations were determined by the DC protein assay (Bio-Rad, Hercules, CA). Proteins were separated by SDS-PAGE, transferred onto polyvinylidene difluoride membranes, and detected with antibodies. Rabbit polyclonal antibodies to rat SUV39H2 were prepared by GeneScript Corporation (Piscataway, NJ). The immunogen consisted of a peptide (CGRKRKAITSKDNNK) corresponding to amino acids 175 to 189 of rat SUV39H2 conjugated to Keyhole Limpet Hemocyanin. SUV39H2 antibodies were affinity-purified prior to use in the western blot experiments. Antibodies to glyceraldehyde 3-phosphate dehydrogenase (GAPDH, 2302, Millipore, Billerica, MA) were also used in the western blot analysis. Immunoreactive proteins were visualized by enhanced chemiluminescence according to the manufacturer’s instructions (Amersham Biosciences, Piscataway, NJ).
2.5. Immunocytochemistry
Immunocytochemical analyses were performed on 10 μm frozen tissue sections. Sections were incubated with 10% normal goat serum (Thermo-Fisher) for 1 h, then overnight with primary antibodies to SUV39H2 (see above) or pan cytokeratin (F3418, Sigma Aldrich). Blastocysts fixed in 4% paraformaldehyde, washed with phosphate buffered saline (PBS, pH 7.4), and permeabilized in PBS containing 0.25% Triton X100. Following a blocking step in 10% normal goat serum for 30 min, blastocysts were incubated with the designated primary antibodies to caudal type homeobox 2 (CDX2, ab76541, Abcam, Cambridge, MA) or POU domain, class 5, transcription factor 1 (POU5F1, also called OCT4, C-10, Santa Cruz Biotechnologies, Santa Cruz, CA) overnight at 4°C. After washing with PBS, sections were incubated for 2 h with corresponding secondary antibodies: Alexa488-conjugated goat-anti-mouse immunoglobulin G (IgG, A32723, Thermo-Fisher) or Alexa 568-conjugated goat-anti-rabbit IgG (A11011, Thermo-Fisher). Nuclei were visualized with 4’,6-diamidino-2-phenylindole (DAPI, Molecular Probes, Carlsbad, CA). Fluorescence images were captured using a Leica DMI 4000 microscope equipped with a charge-coupled device camera (Leica Microsystems GMbH, Welzlar, Germany).
2.6. In situ hybridization
Detection of human SUV39H2 and CGB transcripts was performed on paraffin-embedded human placenta tissue sections using the RNAScope® 2-plex chromogenic assay (Advanced Cell Diagnostics, Hayward, CA), according to the manufacturer’s instructions. Probes were prepared to detect SUV39H2 (454611, accession No. NM_001193424.1) and CGB3 (454831-C2, accession No. NM_000737.3). The CGB3 probe exhibits high homology with CGB5 and CGB8 and is viewed as a generic probe for CGB. Images were captured using a Leica DMI 4000 microscope equipped with a charge-coupled device camera (Leica Microsystems GMbH, Welzlar, Germany).
2.7. Short hairpin RNA (shRNA) constructs and production of lentivirus
pLKO.1 vector harboring Suv39h2 shRNA constructs were obtained from Sigma-Aldrich. Five shRNAs were tested for each gene evaluated. The Suv39h2 shRNA sequences used in this analysis are as follows: i) Suv39h2 shRNA-1: 5’-CCGGCAGCTCGATATGGAAACGTATCTCGAGATACGTTTCCATATCGAGCTGTTTTTG-3’; ii) Suv39h2 shRNA-2: 5’-CCGGTGATACTCTCCAGGAATTATGCTCGAGCATAATTCCTGGAGAGTATCATTTTTG-3’. The Cdx2 shRNA sequences used in the analysis are as follows: i) Cdx2 shRNA-1: 5’-CCGGCGGGTGGTGTACACAGACCATCTCGAGATGGTCTGTGTACACCACCCGTTTTTG-3’; ii) Cdx2 shRNA-2: 5’-CCGGACGTGAGCATGTACCCTAGCTCTCGAGAGCTAGGGTACATGCTCACGTTTTTTG-3’. The control shRNA, which targets no known mammalian gene, pLKO.1-shSCR (Plasmid 1864), was obtained from Addgene (Cambridge, MA) and has the following sequence 5’-CCTAAGGTTAAGTCGCCCTCGCTCTAGCGAGGGCGACTTAACCTTAGG-3’ (30). Lentiviral packaging vectors were obtained from Addgene and included: pMDLg/pRRE (Plasmid 12251), pRSV-Rev (Plasmid 12253) and pMD2.G (Plasmid 12259). Lentiviral particles were produced as previously reported (31, 32). In brief, 293FT cells (Invitrogen) were transiently transfected using Lipofectamine 2000 (Invitrogen) with the following plasmids: shRNA containing transducing vector, third-generation packaging system plasmids (pMDLg/pRRE and pRSV-Rev), and a VSVG envelope plasmid (pMD2.G). The transfection mixture was removed after 8 h and Opti-MEM medium (Invitrogen) supplemented with 5% FBS added. Cell culture supernatants containing lentiviral particles were harvested every 24 h for two days. Supernatants were centrifuged (1,000 x g for 15 min) to remove cell debris, filter (0.45 μm) sterilized, concentrated by ultracentrifugation (35,000 x g for 3h), resuspended in TS Complete culture medium, and stored at −80°C until used for transduction. Lentiviral vector titers were determined by measurement of p24 gag antigen by enzyme-linked immunoassay (Clonetech/Takara Bio, Mountain View, CA). Viral particle (p24) concentrations of 500 ng/ml were used for transduction.
2.8. RNA sequencing (RNA-seq) analysis
Transcriptomic profiles in control shRNA treated TS cells and SUV39H2 shRNA knockdown TS cells (n=3 each) were performed using RNA-seq analysis. Complementary DNA libraries from total RNA samples were prepared with Illumina TruSeq RNA sample preparation kits (Illumina, San Diego, CA). Five hundred ng of total RNA were used as input. Poly-A containing RNAs were purified with oligo-dT-coated magnetic beads. RNA fragmentation, first and second strand cDNA synthesis, end repair, adaptor ligation, and PCR amplification were performed according to the manufacturer’s recommendations. The cDNA libraries were validated for RNA integrity using an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA) before sequencing.
cDNA libraries were clustered onto a TruSeq paired-end flow cell, and sequenced (100 bp paired-end reads) using a TruSeq 200 cycle SBS kit (Illumina). Samples were run on an Illumina HiSeq2000 sequencer and sequenced in parallel with other samples to ensure the data generated for each run were accurately calibrated during analysis. Following generation of sequencing images, the pixel-level raw data collection, image analysis, and base calling were performed by Real Time Analysis software (Illumina). The base call files (*.bcl) were converted to *.qseq files by Illumina’s BCL Converter, and the *.qseq files were subsequently converted to *.fastq files for downstream analysis. Reads from *.fastq files were mapped to the rat reference genome (Ensembl Rnor_5.0.78) using CLC Bio Genomics Workbench 7.0 (CLC Bio, Aarhus, Denmark). The mRNA abundance was expressed in reads per kilobase of exon per million reads mapped (RPKM). Statistical significance was calculated by empirical analysis of digital gene expression in the CLC Bio Genomics Workbench. A corrected false discovery rate (FDR) of 0.05 was used as a cutoff for significant differential expression (control vs Suv39h2 knockdown). Functional patterns of transcript expression were further analyzed using Ingenuity Pathway Analysis (IPA, Qiagen, Redwood City, CA) and Database for Annotation, Visualization and Integrated Discovery (DAVID v6.7; https://david.ncifcrf.gov/). Results from the RNA-seq analysis were validated using RT-qPCR. Primer sets for the RT-qPCR are shown in Table 1.
2.9. Chromatin immunoprecipitation (ChIP)
ChIP analysis was performed according to a previously published procedure (27, 33). Briefly, rat TS cells were grown to confluence in 150-mm dishes and transfected with corresponding shRNAs. Cells were then fixed with 1% formaldehyde, and purified nuclear lysates were sonicated on ice to prepare DNA fragments approximately 500 bp in length. Lysates were immunoprecipitated with 5 μg of antibodies to H3K9me2, H3K9me3, SUV39H2, or CDX2 (see above) and collected on protein A/G-agarose beads (Sigma-Aldrich). Rabbit IgG or mouse IgG (BD Biosciences, Franklin Lakes, NJ) was used as a nonspecific control. Immunoprecipitated chromatin fragments were washed and eluted from protein A-agarose beads. DNA-protein complexes were reverse cross-linked and purified by using a QIAquick PCR purification kit (Qiagen). Purified DNA fragments were characterized by qPCR using primers listed in Table 2. Occupancy/enrichment was normalized to values for input samples by use of the ΔΔCt method and presented relative to values for IgG controls.
Table 2.
Primers used for ChIP analysis.
| Gene | Forward | Reverse |
|---|---|---|
| Adm | CAGACGTGGTGAATTCCTCATCTA | GACTGGCAACTCGGAGAACTTT |
| Cited2 | GCACAAAGGCACGGCTC | CCAACCTGAGAAGAATGGCTATG |
| Foxo4 | GAGTCCGGGACGACCTCTAA | ACCTTGCAATCAGGAGTCCG |
| Suv39h2 | AGTAGCAAACTGAATAGAAGGCATA | AACCAGTTCAGCTGTTATTTCCC |
2.10. Ex vivo lentiviral trophoblast shRNA delivery and analyses
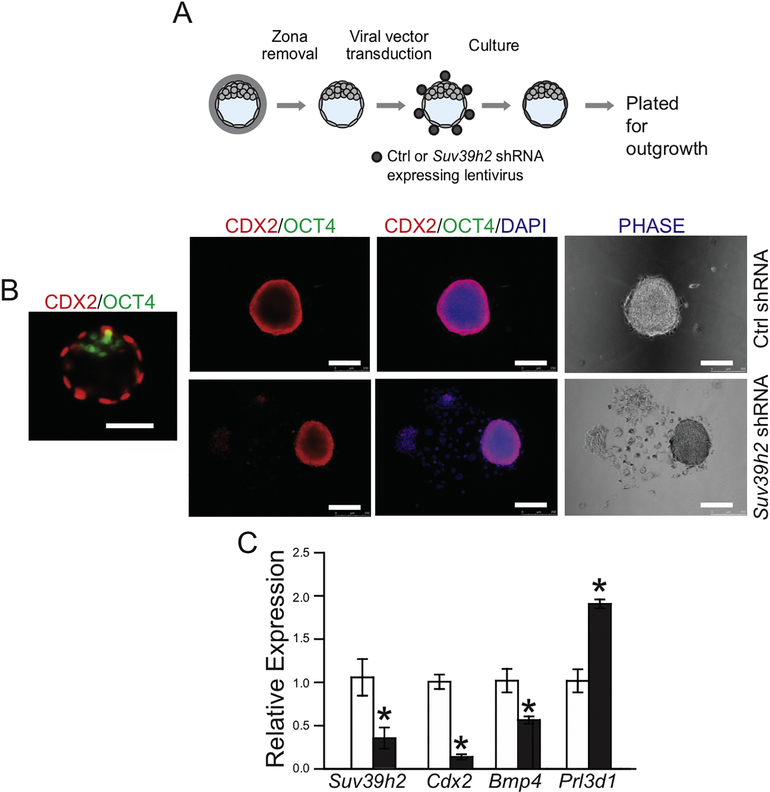
Rat blastocysts were collected by flushing uteri with TS Cell Complete Medium at gd 4.5. Rat blastocysts were transduced with lentiviral particles as previously described (32) containing control or Suv39h2 shRNAs. Briefly, blastocysts were treated with Pronase (Sigma-Aldrich; 10 mg/ml for 10 min) to remove zonae pellucidae and incubated with concentrated lentiviral particles for 4.5 h. Transduced blastocysts were cultured ex vivo in TS Cell Complete Medium. After three days the attached blastocysts were either fixed in 4% paraformaldehyde solution, or collected for RNA isolation (Picopure RNA Isolation Kit, Thermo-Fisher). Blastocyst outgrowth analysis was performed as reported before (27, 32, 34). The surface area of blastocyst outgrowth was visualized using light microscopy and measured using Image J software.
2.11. Statistical analyses
Values are expressed as the mean ± the standard error of the mean (SEM). All experiments were conducted in at least triplicate and were replicated two to three times. Statistical comparisons between two means were evaluated with Student’s t test. Analysis of variance and Tukey’s post hoc tests were used in assessing differences among three or more means using GraphPad Prism (GraphPad Software Inc, La Jolla, CA).
3. Results
3.1. SUV39H2 expression during trophoblast development
TS cells represent a reservoir of cells for trophoblast cell differentiation and organization of the hemochorial placentation site. The TS cell is defined by its transcriptome and its epigenome (5, 35–37), which includes histone H3K9 methylation status (26). Thus far, twelve histone H3K9 methyltransferases have been identified (38).
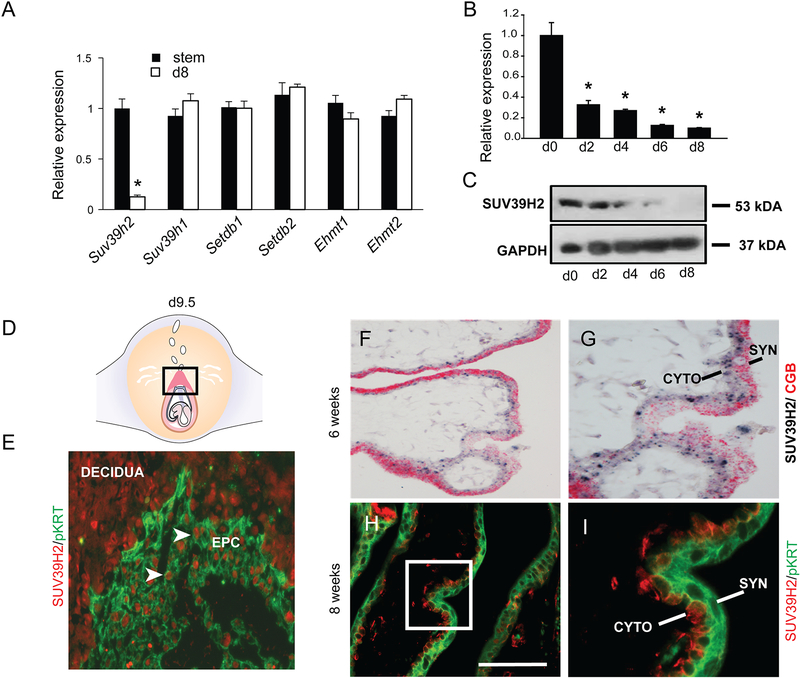
Histone H3K9 methyltransferase gene expression was screened by RT-qPCR in TS cell stem and differentiation states as a first step in defining potential candidates regulating the TS cell epigenetic landscape. Transcripts for members of the SUV39 family of histone H3K9 methyltransferases (Suv39h1, Suv39h2, Ehmt1, Ehmt2, Setdb1, and Setdb2) were readily detectable but only Suv39h2 exhibited a differential pattern of expression in stem and differentiated states (Fig. 1A). Transcripts for other histone H3K9 methyltransferases of the EZH (Kmt2h, Kmt3f), SET2 (Kmt8a, Kmt8e), and PRDM (Kmt8d, Prdm16) families were only detected at extended cycle numbers and were not pursued. SUV39H2 was originally identified as a testis-specific histone H3K9 methyltransferase (39). SUV39H2 transcript and protein were prominently expressed in TS cells and declined as trophoblast cells differentiated (Fig. 1B, C). Consistent with these observations, SUV39H2 protein was detected in locations where stem and progenitor cell populations reside within the developing gd 9.5 rat placenta (ectoplacental cone, Fig. 1D, E). SUV39H2 expression was not restricted to the trophoblast lineage. Expression of SUV39H2 protein was also observed in nuclei of mesometrial decidual cells (Fig. 1E). The significance of SUV39H2 in decidual cells is unknown. The localization of SUV39H2 transcript and protein in were detected in the stem/progenitor cells of the cytotrophoblast layer of the first trimester human placenta (Fig. 1F–I). The syncytiotrophoblast layer, which was identified by CGB transcript and cytokeratin protein expression, was negative for SUV39H2 (Fig. 1F–I). In summary, the SUV39H2 expression profile implicated SUV39H2 as a conserved candidate regulator of TS cells.
Fig. 1. SUV39H2 expression in trophoblast cells.
A) Suv39h2 transcripts exhibit robust differences in stem versus differentiation states (d8) unlike transcripts for other histone H3K9 methyl transferases (Suv39h1, Setdb1, Setdb2, Ehmt1, and Ehmt2), which show modest expression differences during differentiation. SUV39H2 transcript (B) and protein (C) expression is significantly elevated in TS cells and declines as TS cells differentiate. Transcripts were measured by RT-qPCR and proteins assessed by western blotting. D) Schematic of the gd 9.5 conceptus – the boxed area depicts the ectoplacental cone (EPC) of the developing rat placenta. E) SUV39H2 protein immunolocalization to nuclei (red) within stem/progenitor cells of the EPC and surrounding decidua of the gd 9.5 rat conceptus. The EPC is defined as the cellular area positive for pan-cytokeratin (pKRT) (green). F) In situ hybridization co-localization of SUV39H2 (black) and CGB (red) transcripts in first trimester human placenta (6 weeks). G) High magnification of panel F. H) SUV39H2 protein immunolocalization to nuclei (red) within stem/progenitor cells (cytotrophoblast) of the first trimester placenta (8 weeks). I) High magnification of boxed area in panel H. A pan-cytokeratin (pKRT) antibody was used to detect trophoblast cells (green) in both rat and human placental tissues. CYTO, cytotrophoblast; SYN, syncytiotrophoblast. Asterisks indicate significant differences between stem and differentiated cell states, *P<0.01.
3.2. SUV39H2 regulation of TS cells
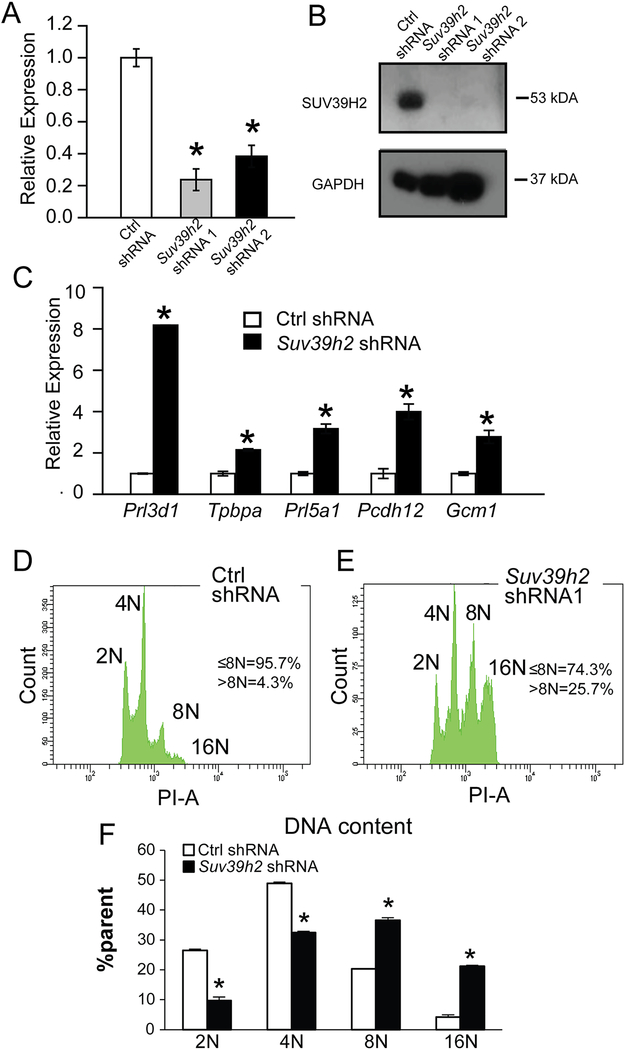
Given the prominence of SUV39H2 in TS cells (Fig. 1), we next investigated its involvement in the regulation of TS cells. SUV39H2 expression was successfully inhibited with lentiviral delivered Suv39h2 shRNAs (Fig. 2A, B). SUV39H2 knockdown TS cells were examined in TS Cell Complete Medium, conditions typically promoting the stem state. Disruption of SUV39H2 expression interfered with cell proliferation and led to phenotypic changes indicative of cell differentiation, including flow cytometric evidence of endoreduplication (Fig. 2C, D) and the significant upregulation of transcripts characteristic of multi-lineage trophoblast cell specializations (32, trophoblast giant cell, Prl3d1; spongiotrophoblast, Tpbpa; invasive trophoblast, Prl5a1; glycogen cell, Pcdh12; syncytial trophoblast, Gcm1; Fig. 2E). These observations prompted a more extensive analysis of the effect of SUV39H2 knockdown on the TS cell phenotype.
Fig. 2. Disruption of SUV39H2 expression has striking effects on TS cells.
Validation of shRNA knockdown of SUV39H2 transcript by RT-qPCR (A) and protein by western blotting (B) in TS cells. C) Knockdown of SUV39H2 was associated with an upregulation of indices of trophoblast giant cell (Prl3d1), spongiotrophoblast (Tpbpa), invasive trophoblast (Prl5a1), glycogen trophoblast (Pcdh12), and syncytiotrophoblast (Gcm1) differentiation. D-F) SUV39H2 knockdown inhibited TS cell proliferation and promoted trophoblast giant cell formation as assessed by flow cytometry of propidium stained nuclei. Asterisks indicate significant differences between control and SUV39H2 knockdown cells, *P<0.01.
3.3. Impact of SUV39H2 on the TS cell transcriptome
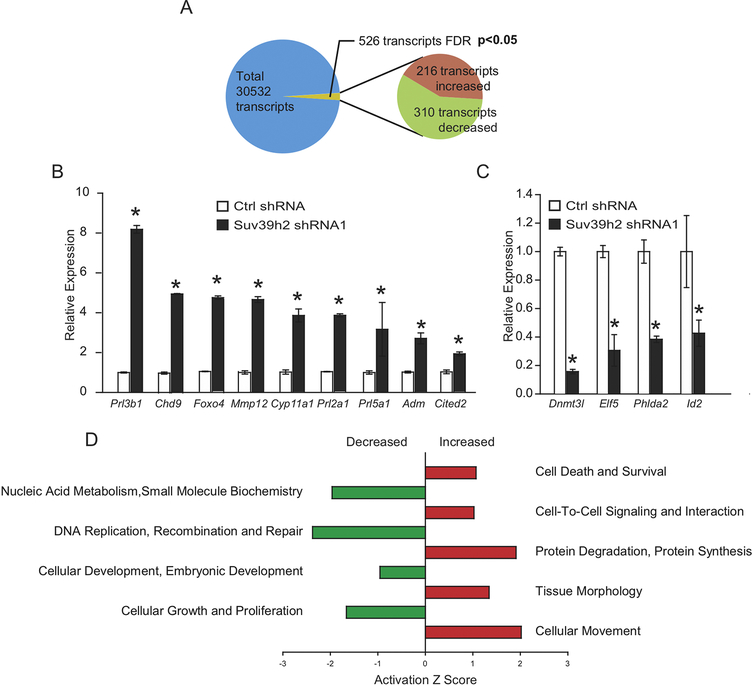
RNA-seq was performed in TS cells under control stem state conditions or following SUV39H2 knockdown. A total of 526 transcripts were significantly affected by SUV39H2 disruption, including 310 downregulated transcripts and 216 upregulated transcripts (false discovery rate, P<0.05; Fig. 3A, Supplementary Table 1). Decreases in transcript expression were prominently associated with cellular processes such as: proliferation and DNA replication, whereas increases in transcript expression were associated with cellular processes such as: movement, protein synthesis, and cell-cell signaling (Fig. 3B). Subsets of the SUV39H2-dependent transcripts were selected for further analyses by RT-qPCR. Representative downregulated transcripts, included those associated with maintaining TS cells (Dnmt3l, Phlda2, Elf5, and Id2; Fig. 3C), while upregulated transcripts included those associated with the differentiation state (Prl3b1, Adm, Mmp12, Cyp11a1, and Prl2a1; Fig. 3D), and potential mediators of differentiation (Foxo4, Cited2, and Chd9; Fig. 3E). The findings indicate that SUV39H2 contributes to maintenance of TS cells and an inhibition of trophoblast cell differentiation.
Fig. 3. Effects of SUV39H2 on the TS cell transcriptome.
A) Overall, 526 transcripts were significantly different between control and SUV39H2 knockdown TS cells (P<0.05). Of the differentially regulated transcripts, 216 transcripts were upregulated, and 310 transcripts were downregulated in the SUV39H2 knockdown TS cells. B) RT-qPCR validation of select upregulated transcripts. C) RT-qPCR validation of select downregulated transcripts. D) Pathway analysis of transcripts differentially affected by SUV39H2 disruption (Ingenuity Pathway Analysis). Asterisks indicate significant differences between control and SUV39H2 knockdown cells, *P<0.01.
3.4. SUV39H2 and the histone H3K9 epigenetic landscape
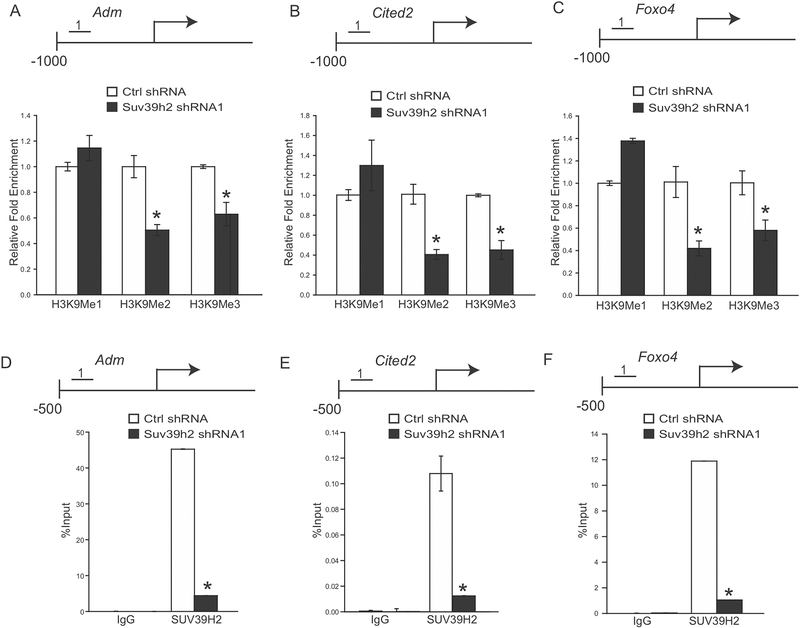
SUV39H2 is a lysine methyltransferase with known actions on dimethylation and trimethylation of histone H3K9 (38–40). shRNA mediated knockdown of SUV39H2 resulted in upregulation of a several transcripts previously implicated in trophoblast cell differentiation. We hypothesized that SUV39H2 regulates the H3K9 methylation landscape of its target genes. H3K9 methylation status of putative SUV39H2 target genes (Adm, Foxo4, and Cited2) was investigated by ChIP qPCR analysis. Dramatic changes in H3K9 methylation were observed within the proximal promoter regulatory regions for Adm, Foxo4, and Cited2 genes. Unlike H3K9me1, H3K9me2 and H3K9me3 modifications were sensitive to SUV39H2 knockdown (Fig. 4A–C). ChIP analyses also indicated that SUV39H2 occupied these targets sites, suggesting potentially direct action at the affected loci.
Fig. 4. Histone H3K9 methylation landscape associated with SUV39H2 targets in TS cells.
A) Schematic layout of the rat Adm gene and the location of one of the regions (No. 1: −807 to −659 bp upstream of the transcription start site) surveyed in control and SUV39H2 knockdown TS cells by chromatin immunoprecipitation (ChIP) analysis for H3K9me1, H3K9me2, and H3K9me3. B) Schematic layout of the rat Cited2 gene and the location of one of the regions (No. 1: −537 to −431 bp upstream of the transcription start site) surveyed in control and SUV39H2 knockdown TS cells by ChIP analysis for H3K9me1, H3K9me2, and H3K9me3. C) Schematic layout of the rat Foxo4 gene and the location of one of the regions (No. 1: −214 to −117 bp upstream of the transcription start site) surveyed in control and SUV39H2 knockdown TS cells by ChIP analysis for H3K9me1, H3K9me2, and H3K9me3. D) ChIP analysis with IgG or antibodies specific to SUV39H2 at the Adm locus in control and SUV39H2 knockdown TS cells. E) ChIP analysis with IgG or antibodies specific to SUV39H2 at the Cited2 locus in control and SUV39H2 knockdown TS cells. F) ChIP analysis with IgG or antibodies specific to SUV39H2 at the Foxo4 locus in control and SUV39H2 knockdown TS cells. Asterisks indicate significant differences between control and SUV39H2 knockdown cells, *P<0.01.
3.5. Ex vivo analysis of SUV39H2 in blastocyst-derived TS cell expansion
In an effort to examine the role of SUV39H2 in an ex vivo setting we investigated the effect of SUV39H2 knockdown on trophoblast cell behavior in cultured blastocysts (Fig. 5A). CDX2 exhibits specific expression in the outer trophectoderm cell layer, while OCT4 (POU5F1) demarcates the inner cell mass (Fig. 5B). Culture of control blastocysts in conditions promoting TS cell expansion was associated with the establishment of tightly packed CDX2 positive colonies and minimal outgrowth of differentiated trophoblast cells (Fig. 5B). In contrast, disruption of SUV39H2 resulted in extensive outgrowth of differentiated trophoblast cells (Fig. 5B). Furthermore, knockdown of SUV39H2 diminished expression of Suv39h2 and other indices of the TS stem cell state (Cdx2, and Bmp4) and a corresponding upregulation of Prl3d1 expression, a marker of trophoblast giant cell differentiation (Fig. 5C). Thus, SUV39H2 exhibits a parallel pattern of regulatory activities in TS cells and in blastocysts.
Fig. 5. Ex vivo analyses of SUV39H2-specific shRNA treated blastocysts.
A) Schematic showing the experimental plan for lentiviral transduction of blastocysts and outgrowth assay. Blastocysts were transduced with control (Ctrl) or Suv39h2 shRNA and examined following culture. B) Representative images of a blastocyst immunostained for CDX2 and OCT4 and blastocyst outgrowths immunostained for CDX2 and OCT4 (left panels) and stained for DAPI (middle panels) or photographed under phase microscopy (right panels) from: Ctrl shRNA and Suv39h2 shRNAs, cultured in stem state culture conditions. C) Suv39h2, Cdx2, Bmp4, and Prl3d1 transcripts in control and knockdown cultures were measured by RT-qPCR. Asterisks indicate significant differences between control and SUV39H2 knockdown blastocyst cultures, *P<0.01.
3.6. CDX2 is an upstream regulator of TS cell SUV39H2
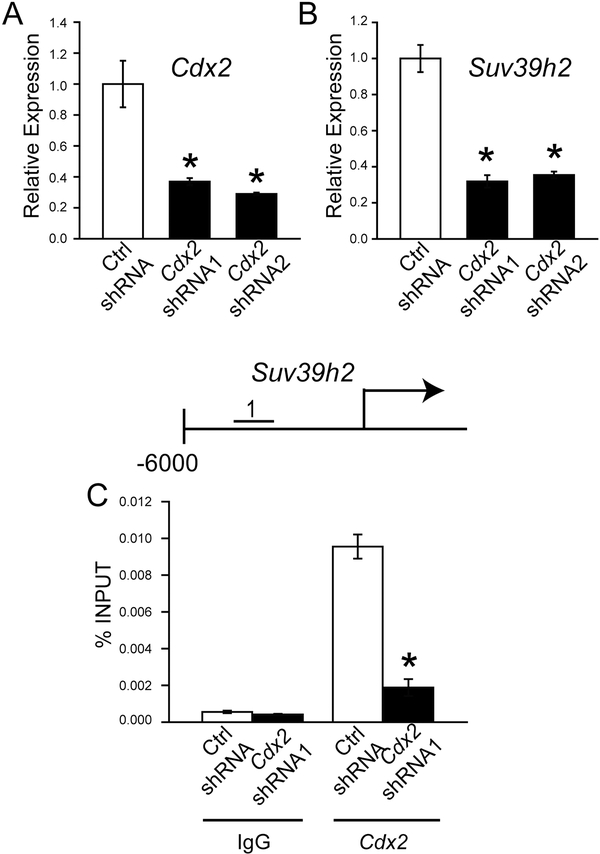
Several transcriptional regulators of TS cells have been identified (3, 6, 41). Among these transcriptional regulators, CDX2 shows a pivotal role in establishing the trophoblast cell lineage (42–44). Consequently, we examined the connection between CDX2 and SUV39H2. Inhibition of Cdx2 expression using CDX2-specific shRNAs led to a significant disruption in Suv39h2 expression (Fig. 6A and B). Furthermore, ChIP analysis demonstrated that CDX2 accumulates at the 5’ regulatory region of the Suv39h2 locus. The specificity of the interaction between CDX2 and the Suv39h2 locus was demonstrated in CDX2 shRNA expressing TS cells. This experimentation is consistent with CDX2 possessing an upstream regulatory role controlling SUV39H2.
Fig. 6. Hierarchal regulation of SUV39H2 in TS cells.
A) Validation of Cdx2 shRNA knockdown of Cdx2 transcript by RT-qPCR. B) Effects of Cdx2 shRNA knockdown on Suv39h2 transcript by RT-qPCR. C) Schematic layout of the rat Suv39h2 gene and the location of one of the regions (No. 1: 4369 to −4200 bp upstream of the transcription start site) surveyed in control and CDX2 knockdown TS cells by ChIP analysis with IgG or antibodies specific to CDX2. Asterisks indicate significant differences between control and CDX2 knockdown cells, *P<0.01.
In summary, our findings indicate that SUV39H2 contributes to the maintenance of TS cells and restrains trophoblast cell differentiation and thus serves as a contributor to the epigenetic regulation of the trophoblast cell lineage and placental development.
4. Discussion
The placenta governs fetal development. Specialized trophoblast cells arise from stem/progenitor cell populations with the capacity to facilitate delivery of nutrients from the mother to the placenta or from the placenta to the fetus (45, 46). Thus, maintenance, expansion, and ultimately differentiation of TS cells are pivotal events in placentation. The regulation of these events ultimately determines the functionality of the placenta. In this report, we identified SUV39H2 as a regulator of TS cell development. SUV39H2 expression is elevated in the stem state and declines following TS cell differentiation. The TS cell state is characterized by self-renewal and a restraint on trophoblast cell differentiation, cellular processes shown to be regulated by SUV39H2.
SUV39H2 modulates the expression of genes pivotal to the maintenance of TS cells and characteristic of the differentiation state. Genes required for maintaining TS cells in the stem state, including Dnmt3l, Elf5, Phlda2, and Id2 (47–53), are dependent on SUV39H2 for optimal expression. Simultaneously, SUV39H2 expression in the stem state inhibits trophoblast cell differentiation. Interference of SUV39H2 expression results in the formation of trophoblast giant cells and the upregulation of transcripts associated with multiple lineages of differentiated trophoblast cells (29, 32, 54, 55). Thus, SUV39H2 plays a pivotal decision-making role dictating trophoblast cell contributions to the placentation process.
SUV39H2 is a methyltransferase (39). Among the H3K9 methyltransferases, SUV39H2 was the only methyltransferase to exhibit a differential expression pattern based on the trophoblast cell differentiation state. One of the outcomes of H3K9 methylation is transcriptional repression (10). SUV39H2 was directly linked to H3K9 methylation within regulatory regions of three genes activated during differentiation, Foxo4, Cited2, and Adm. FOXO4 is a member of the forkhead family of transcription factors (56) and is a downstream effector of the phosphatidylinositol-3 kinase/AKT signaling pathway (57), which has been shown to regulate trophoblast cell differentiation (55, 58–61). CITED2 is a transcriptional co-regulator as exemplified in its ability to modulate Cbp/p300 recruitment to members of the hypoxia inducible factor and activator protein-2 transcription factor families (62). CITED2 is also a known regulator of placentation (63, 64). ADM is a ligand produced by trophoblast giant cells with a spectrum of actions on uterine vascular and immune cells (65–68). The actions of SUV39H2 on Foxo4, Cited2, and Adm are consistent with a transcriptionally repressive role for H3K9 methylation. Rugg-Gunn and colleagues have also established a regulatory role for H3K9 methylation in early trophoblast development (26). The mode of SUV39H2 action in modulating TS cell-associated genes is not known but may be linked to its actions on lysine-specific demethylase 1 (LSD1, also known as lysine demethylase 1A, KDM1A). LSD1 is a target of SUV39H2 methyltransferase actions (69). SUV39H2-mediated methylation stabilizes the LSD1 protein. LSD1 is an H3K4 demethylase (70) critically important in maintaining TS cells (71–73). Although, collectively these observations indicate a pivotal regulatory role for SUV39H2 in maintaining TS cells, they do not preclude other H3K9 methyltransferases, including those showing similar expression profiles in stem and differentiating TS cells, from having a role in regulating the trophoblast cell epigenetic landscape and dictating TS cell development.
SUV39H2 is situated downstream of CDX2 in the regulatory hierarchy controlling trophoblast cell development. CDX2 and SUV39H2 exhibit a similar pattern of expression in TS cells: elevated in the stem state and diminished following TS cell differentiation (29, present study). SUV39H2 expression was critically dependent on CDX2 expression. Furthermore, accumulation of CDX2 at the Suv39h2 locus is consistent with CDX2 directly regulating Suv39h2 gene transcription. Disruption of Cdx2 impairs trophoblast cell development resulting in lethality during early pregnancy (74, 75). In contrast, Suv39h2 inactivation in the mouse does not phenocopy the Cdx mutant phenotype and was compatible with prenatal development (76). Placental development was not investigated in Suv39h2 mutants. Thus, a sublethal placental phenotype may have been exhibited or alternatively, other H3K9 methyltransferases may have compensated for the in vivo absence of SUV39H2. Of note, mice possessing null mutations of both Suv39h1 and Suv39h2 exhibit severe growth restriction and impaired postnatal viability (76).
In conclusion, SUV39H2 is an epigenetic regulator contributing to the modulation of TS cells, impacting TS cell expansion and trophoblast cell differentiation, essential events in the development of the hemochorial placenta.
Supplementary Material
Highlights.
SUV39H2 is preferentially expressed in trophoblast stem cells
SUV39H2 promotes maintenance of trophoblast stem cells and restrains differentiation
CDX2 is an upstream regulator of SUV39H2 in trophoblast stem cells
SUV39H2 contributes to the epigenetic landscape of trophoblast stem cells
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (HD020676, HD079363, HD099638) and the Sosland Foundation. We express our sincere appreciation to the late Dr. Shui Qing Ye for assistance with the RNA-seq analysis. We thank Stacy Oxley and Brandi Miller for administrative assistance.
Footnotes
Data availability
The RNA-seq dataset is available in the Gene Expression Omnibus website (https://www.ncbi.nlm.nih.gov/geo/; accession no. GSE154701). The remainder of the data is contained within the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cockburn K and Rossant J (2010) Making the blastocyst: lessons from the mouse. J. Clin. Invest. 120, 995–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cha J, Sun X, and Dey SK (2012) Mechanisms of implantation: strategies for successful pregnancy. Nat. Med. 18, 1754–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfeffer PL and Pearton DJ (2012) Trophoblast development. Reproduction 143, 231–246 [DOI] [PubMed] [Google Scholar]
- 4.Roberts RM and Fisher SJ (2011) Trophoblast stem cells. Biol. Reprod. 84, 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rugg-Gunn PJ (2012) Epigenetic features of the mouse trophoblast. Reprod. BioMed. Online 25, 21–30 [DOI] [PubMed] [Google Scholar]
- 6.Latos PA and Hemberger M (2016) From the stem of the placental tree: trophoblast stem cells and their progeny. Development 143, 3650–3660 [DOI] [PubMed] [Google Scholar]
- 7.Koziol MJ and Rinn JL (2010) RNA traffic control of chromatin complexes. Curr. Opin. Genet. Dev. 20, 142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santos F, Peters AH, Otte AP, Reik W, and Dean W (2005) Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev. Biol. 280, 225–236 [DOI] [PubMed] [Google Scholar]
- 9.Burton A and Torres-Padilla ME (2014) Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat. Rev. Mol. Cell. Biol. 15, 723–734 [DOI] [PubMed] [Google Scholar]
- 10.Kouzarides T (2007) Chromatin modifications and their function. Cell 21, 381–395 [DOI] [PubMed] [Google Scholar]
- 11.Ko SY, Kang Y, Lee HS, Han SY, and Hong SH. (2006) Identification of Jmjd1a as a STAT3 downstream gene in mES cells. Cell Struct. Funct. 31, 53–62 [DOI] [PubMed] [Google Scholar]
- 12.Lockman K, Taylor JM, and Mack CP (2007) The histone demethylase, Jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ. Res. 101, e115–e123 [DOI] [PubMed] [Google Scholar]
- 13.Loh Y-H, Zhang W, Chen X, George J, and Ng H-H (2007) Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 21, 2545–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen B, Wu H, Shinkai Y, Irizarry RA, and Feinberg AP (2009) Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 41, 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi HH, Sarkissian M, Hu G-Q, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, Yaghi NK, Lim H, Garcia BA, Brizuela L, Zhao K, Roberts TM, and Shi Y (2010) Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466, 503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strobl-Mazzulla PH, Sauka-Spengler T, and Bronner-Fraser M (2010) Histone demethylase JmjD2A regulates neural crest specification. Dev. Cell 19, 460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawazu M, Saso K, Tong KI, McQuire T, Goto K, Son D-O, Wakeham A, Miyagishi M, Mak TW, and Okada H (2011) Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS ONE 6, e17830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi L, Sun L, Li Q, Liang J, Yu W, Yi X, Yang X, Li Y, Han X, Zhang Y, Xuan C, Yao Z, Shang Y 2011. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc. Natl. Acad. Sci. USA 108, 7451–7456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verrier L, Escaffit F, Chailleux C, Trouche D, and Vandromme M (2011) A New Isoform of the histone demethylase JMJD2A/KDM4A is required for skeletal muscle differentiation. PLoS Genet. 7, e1001390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herzog M, Josseaux E, Dedeurwaerder S, Calonne E, Wolkmar M, and Fuks F 2012. The histone demethylase Kdm3a is essential to progression through differentiation. Nucleic Acids Res. 40, 7219–7232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, Yaw LP, Robson P, Lim B, Ng HH (2009) Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 23, 2507–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeap LS, Hayashi K, and Surani MA (2009) ERG-associated protein with SET domain (ESET)-Oct4 interaction regulates pluripotency and represses the trophectoderm lineage. Epigenetics Chromatin 2, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohmann F, Loureiro J, Su H, Fang Q, Lei H, Lewis T, Yang Y, Labow M, Li E, Chen T, and Kadam S (2010) KMT1E mediated H3K9 methylation is required for the maintenance of embryonic stem cells by repressing trophectoderm differentiation. Stem Cells 28, 201–212 [DOI] [PubMed] [Google Scholar]
- 24.Alder O, Lavial F, Helness A, Brookes E, Pinho S, Chandrashekran A, Arnaud P, Pombo A, O’Neill L, and Azuara V (2010) Ring1b and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment. Development 137, 2483–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park K-E, Johnson CM, Wang X, and Cabot RA (2011) Differential developmental requirements for individual histone H3K9 methyltransferases in cleavage-stage porcine embryos. Reprod. Fertil. Dev. 23, 551–560 [DOI] [PubMed] [Google Scholar]
- 26.Rugg-Gunn PJ, Cox BJ, Ralston A, and Rossant J (2010) Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc. Natl. Acad. Sci. USA 107, 10783–10790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakraborty D, Cui W, Rosario GX, Scott RL, Dhakal P, Renaud SJ, Tachibana M, Rumi MAK, Mason CW, Krieg AJ, and Soares MJ (2016) HIF-KDM3A-MMP12 regulatory circuit ensures trophoblast plasticity and placental adaptations to hypoxia. Proc. Natl. Acad. Sci. U.S.A. 113, E7212–E7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ain R, Konno T, Canham LN, and Soares MJ (2006) Phenotypic analysis of the rat placenta. Methods Mol. Med. 121, 295–313 [DOI] [PubMed] [Google Scholar]
- 29.Asanoma K, Rumi MAK, Kent LN, Chakraborty D, Renaud SJ, Wake N, Lee D-S, Kubota K, and Soares MJ (2011) FGF4-dependent stem cells derived from rat blastocysts differentiate along the trophoblast lineage. Dev. Biol. 351, 110–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarbassov DD, Guertin DA, Ali SM, and Sabatini DM (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 31.Lee DS, Rumi MA, Konno T, and Soares MJ (2009) In vivo genetic manipulation of the rat trophoblast cell lineage using lentiviral vector delivery. Genesis 47, 433–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakraborty D, Muto M, and Soares MJ (2017) Ex vivo trophoblast-specific genetic manipulation using lentiviral delivery. Bio. Protoc. 7, e2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubota K, Kent LN, Rumi MAK, Roby KF, and Soares MJ (2015) Dynamic regulation of AP-1 transcriptional complexes directs trophoblast differentiation. Mol. Cell. Biol. 35, 3163–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ain R, Canham LN, and Soares MJ (2003) Gestation stage-dependent intrauterine trophoblast cell invasion in the rat and mouse: novel endocrine phenotype and regulation. Dev. Biol. 260, 176–190 [DOI] [PubMed] [Google Scholar]
- 35.Chuong E, Rumi MAK, Soares MJ, and Baker J (2013) Placental endogenous retroviruses facilitate rapid evolution of core trophoblast regulatory network. Nat. Genetics 45, 325–32923396136 [Google Scholar]
- 36.Hemberger M, Dean W, and Reik W (2009) Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat. Rev. Mol. Cell Biol. 10, 526–537 [DOI] [PubMed] [Google Scholar]
- 37.Paul S and Knott JG (2014) Epigenetic control of cell fate in mouse blastocysts: the role of covalent histone modifications and chromatin remodeling. Mol. Reprod. Dev. 81, 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mozzetta C, Boyarchuk E, Pontis J, and Ait-Si-Ali S (2015) Sound of silence: the properties and functions of repressive Lys methyltransferases. Nat. Rev. Mol. Cell Biol. 16, 499–513 [DOI] [PubMed] [Google Scholar]
- 39.O’Carroll D, Scherthan H, Peters AHFM, Opravil S, Haynes AR, Laible G, Rea S, Schmid M, Lebersorger A, Jerratsch M, Sattler L, Mattei MG, Denny P, Brown SDM, Schwizer D, and Jenuwein T (2000) Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol. 20, 9423–9433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice JC, Briggs SD, Ueberhelde B, Barber CM, Shabanowitz J, Hunt CD, Shinkai Y, and Allis CD (2003) Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12, 1591–1598 [DOI] [PubMed] [Google Scholar]
- 41.Hemberger M, Hanna CW, and Dean W (2020) Mechanisms of early placental development in mouse and humans. Nat. Rev. Genet. 21, 27–43. [DOI] [PubMed] [Google Scholar]
- 42.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, and Rossant J. (2005) Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917–929. [DOI] [PubMed] [Google Scholar]
- 43.Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, and Rossant J (2005) Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102. [DOI] [PubMed] [Google Scholar]
- 44.Frum T and Ralston A (2015) Cell signaling and transcription factors regulating cell fate during formation of the mouse blastocyst. Trends Genet. 31, 402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soares MJ, Varberg KM, and Iqbal K (2018) Hemochorial placentation: development, function, and adaptations. Biol. Reprod. 99, 196–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knöfler M, Haider S, Saleh L, Pollheimer J, Gamage TKJB, and James J (2019) Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell Mol. Life Sci. 76, :3479–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arima T, Hata K, Tanaka S, Kusumi M, Li E, Kato K, Shiota K, Sasaki H, and Wake N (2006) Loss of the maternal imprint in Dnmt3Lmat−/− mice leads to a differentiation defect in the extraembryonic tissue. Dev. Biol. 297, 361–373. [DOI] [PubMed] [Google Scholar]
- 48.Donnison M, Beaton A, Davey HW, Broadhurst R, L’Huillier P, and Pfeffer PL (2005) Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development 132, 2299–2308. [DOI] [PubMed] [Google Scholar]
- 49.Pearton DJ, Smith CS, Redgate E, van Leeuwen J, Donnison M, and Pfeffer PL (2014) Elf5 counteracts precocious trophoblast differentiation by maintaining Sox2 and 3 and inhibiting Hand1 expression. Dev. Biol. 392, 344–357. [DOI] [PubMed] [Google Scholar]
- 50.Latos PA, Sienerth AR, Murray A, Senner CE, Muto M, Ikawa M, Oxley D, Burge S, Cox BJ, and Hemberger M (2015) Elf5-centered transcription factor hub controls trophoblast stem cell self-renewal and differentiation through stoichiometry-sensitive shifts in target gene networks. Genes Dev. 29, 2435–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takao T, Asanoma K, Tsunematsu R, Kato K, and Wake N (2012) The maternally expressed gene Tssc3 regulates the expression of MASH2 transcription factor in mouse trophoblast stem cells through the AKT-Sp1 signaling pathway. J. Biol. Chem. 287, 42685–42694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janatpour MJ, McMaster MT, Genbacev O, Zhou Y, Dong J, Cross JC, Israel MA, and Fisher SJ (2000) Id-2 regulates critical aspects of human cytotrophoblast differentiation, invasion and migration. Development 127, 549–558.10631176 [Google Scholar]
- 53.Guo G, Huss M, Tong GQ, Wang C, Sun LL, Clarke ND, and Robson P (2010) Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell. 18, 675–685. [DOI] [PubMed] [Google Scholar]
- 54.Simmons DG and Cross JC (2005) Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev. Biol. 284,12–24. [DOI] [PubMed] [Google Scholar]
- 55.Kent LN, Konno T, and Soares MJ (2010) Phosphatidylinositol 3 kinase modulation of trophoblast cell differentiation. BMC Dev. Biol. 10, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benayoun BA, Caburet S, and Veitia RA (2011) Forkhead transcription factors: key players in health and disease. Trends Genet. 27, 224–232. [DOI] [PubMed] [Google Scholar]
- 57.Link W (2019) Introduction to FOXO biology. Methods Mol. Biol. 1890, 1–9. [DOI] [PubMed] [Google Scholar]
- 58.Kamei T, Jones SR, Chapman BM, McGonigle KL, Dai G, and Soares MJ (2002) The phosphatidylinositol 3-kinase/Akt signaling pathway modulates the endocrine differentiation of trophoblast cells. Mol. Endocrinol. 16, 1469–1481. [DOI] [PubMed] [Google Scholar]
- 59.Yang ZZ, Tschopp O, Hemmings-Mieszczak M, Feng J, Brodbeck D, Perentes E, and Hemmings BA. (2003) Protein kinase B alpha/Akt1 regulates placental development and fetal growth. J. Biol. Chem. 278, 32124–32131. [DOI] [PubMed] [Google Scholar]
- 60.Kent LN, Rumi MAK, Kubota K, Lee D-S, and Soares MJ (2011) FOSL1 is integral to establishing the maternal-fetal interface. Mol. Cell. Biol. 31, 4801–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kent LN, Ohboshi S, and Soares MJ (2012) Akt1 and insulin-like growth factor 2 (Igf2) regulate placentation and fetal/postnatal development. Int. J. Dev. Biol. 56, 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Braganca J, Mendes-Silva L, Lopes JA, and Calado SM (2019) CITED proteins in the heart of pluripotent cells and in heart’s full potential. Regen. Med. Front. 1, e190005. [Google Scholar]
- 63.Withington SL, Scott AN, Saunders DN, Lopes Floro K, Preis JI, Michalicek J, Maclean K, Sparrow DB, Barbera JP, and Dunwoodie SL (2006) Loss of Cited2 affects trophoblast formation and vascularization of the mouse placenta. Dev. Biol. 294, 67–82. [DOI] [PubMed] [Google Scholar]
- 64.Moreau JL, Artap ST, Shi H, Chapman G, Leone G, Sparrow DB, and Dunwoodie SL (2014) Cited2 is required in trophoblasts for correct placental capillary patterning. Dev. Biol. 392, 62–79. [DOI] [PubMed] [Google Scholar]
- 65.Montuenga LM, Martínez A, Miller MJ, Unsworth EJ, and Cuttitta F (1997) Expression of adrenomedullin and its receptor during embryogenesis suggests autocrine or paracrine modes of action. Endocrinology 138, 440–451. [DOI] [PubMed] [Google Scholar]
- 66.Yotsumoto S, Shimada T, Cui CY, Nakashima H, Fujiwara H, and Ko MS (1998) Expression of adrenomedullin, a hypotensive peptide, in the trophoblast giant cells at the embryo implantation site in mouse. Dev. Biol. 203, 264–275. [DOI] [PubMed] [Google Scholar]
- 67.Lenhart PM and Caron KM (2012) Adrenomedullin and pregnancy: perspectives from animal models to humans. Trends Endocrinol. Metab. 23, 524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li M, Schwerbrock NM, Lenhart PM, Fritz-Six KL, Kadmiel M, Christine KS, Kraus DM, Espenschied ST, Willcockson HH, Mack CP, and Caron KM (2013) Fetal-derived adrenomedullin mediates the innate immune milieu of the placenta. J. Clin. Invest. 123, 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Piao L, Suzuki T, Dohmae N, Nakamura Y, and Hamamoto R (2015) SUV39H2 methylates and stabilizes LSD1 by inhibiting polyubiquitination in human cancer cells. Oncotarget 6, 16939–16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi Y (2007) Histone lysine demethylases: emerging roles in development, physiology and disease. Nat. Rev. Genet. 8, 829–833. [DOI] [PubMed] [Google Scholar]
- 71.Zhu D, Hölz S, Metzger E, Pavlovic M, Jandausch A, Jilg C, Galgoczy P, Herz C, Moser M, Metzger D, Günther T, Arnold SJ, and Schüle R (2014) Lysine-specific demethylase 1 regulates differentiation onset and migration of trophoblast stem cells. Nat. Commun. 5, 3174. [DOI] [PubMed] [Google Scholar]
- 72.Latos PA, Goncalves A, Oxley D, Mohammed H, Turro E, and Hemberger M (2015) Fgf and Esrrb integrate epigenetic and transcriptional networks that regulate self-renewal of trophoblast stem cells. Nat. Commun. 6, 7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Castex J, Willmann D, Kanouni T, Arrigoni L, Li Y, Friedrich M, Schleicher M, Wöhrle S, Pearson M, Kraut N, Méret M, Manke T, Metzger E, Schüle R, and Günther T (2017) Inactivation of Lsd1 triggers senescence in trophoblast stem cells by induction of Sirt4. Cell Death Dis. 8, e2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chawengsaksophak K, de Graaff W, Rossant J, Deschamps J, and Beck F (2004) Cdx2 is essential for axial elongation in mouse development. Proc. Natl. Acad. Sci. USA. 101, 7641–7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, and Rossant J (2005) Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102. [DOI] [PubMed] [Google Scholar]
- 76.Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, and Jenuwein T (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.