Figure 4.
PAF positively modulates global cell-cycle genes regulated by the DREAM complex
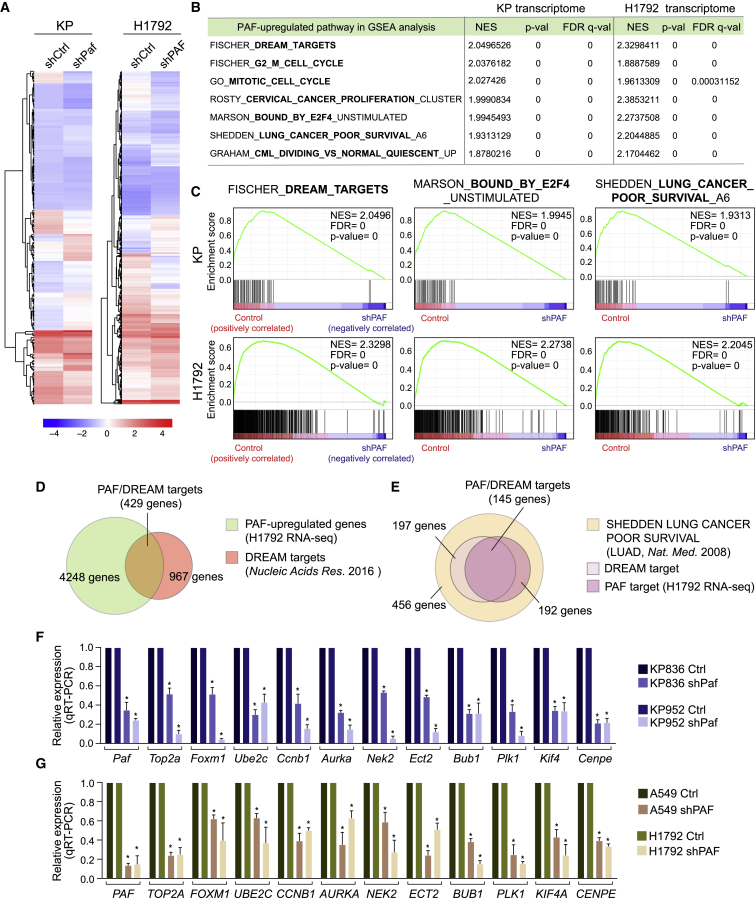
(A) Hierarchically clustered heatmaps of differentially expressed genes (DEGs) in control versus PAF-depleted mouse (KP) and human (H1792) lung cancer cells. DEGs were analyzed by RNA-seq. Mean values are shown (n = 2).
(B) PAF-upregulated pathways in GSEA. Representative cell-cycle- and cancer-related pathways that were highly enriched (high normalized enrichment score [NES] with the lowest p values) and overlapped with PAF-upregulated pathways in mouse (KP) and human (H1792) lung cancer cell lines are shown. Complete GSEA results are in Table S3.
(C) GSEA plots for the DREAM_TARGETS, BOUND_BY_E2F4, and LUNG_CANCER_POOR_SURVIVAL pathways which were upregulated by PAF in mouse KP and human lung cancer H1792 cell lines. The Fischer_DREAM_TARGETS (gene set size, n = 967) gene sets were highly associated with PAF-upregulated genes.
(D) Venn diagram showing overlap of PAF and DREAM complex target genes. PAF upregulated a considerable number (n = 429 of 967, H1792 cells) of DREAM target genes.
(E) Venn diagram showing overlap of PAF-DREAM targets and genes related to poor lung cancer survival.
(F and G) Validation of expression of PAF-DREAM complex-regulated genes in control versus (F) PAF-KD mouse (KP836 and KP952) and (G) human (A549 and H1792) lung cancer cell lines; qRT-PCR assays; error bars indicate SEM; ∗p < 0.05.