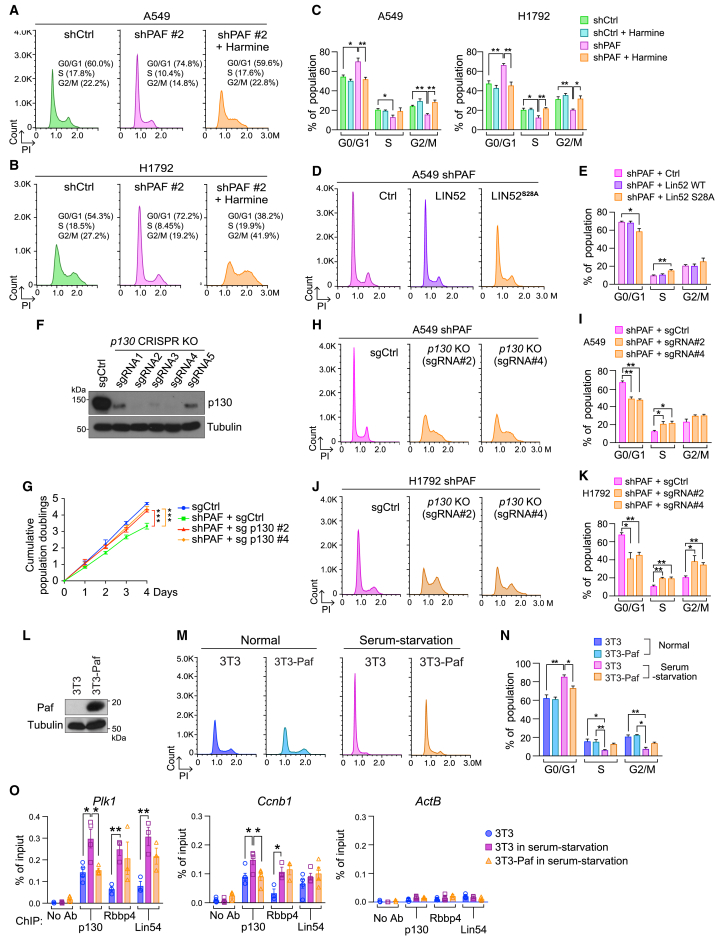
Figure 5.
PAF leads to cell quiescence exit and cell proliferation through repression of the DREAM complex
(A–C) Harmine treatment rescues the G0/G1 arrest of PAF-depleted A549 (A) and H1792 (B) cells. (C) quantifications of cell-cycle distribution shown in (A), (B), and Figure S5C. The cell-cycle distribution of each cell line was analyzed using PI staining and FACS.
(D and E) Ectopic expression of LIN52S28A mutant that no longer binds to p130 in a dominant-negative manner rescues the G0/G1 arrest of PAF-KD A549 cells. (D) The cell-cycle phases in the indicated conditions were analyzed using FACS with PI staining. LIN52 wild-type (WT) cells served as a control group. (E) Quantification of cell-cycle phases.
(F–K) p130 KO rescues the G0/G1 and growth arrest induced by PAF KD. (F) Immunoblotting validation of p130 KO in A549 cells targeted by the CRISPR-Cas9 system (five sgRNAs were used). (G) Cumulative population doublings of PAF-KD A549 cells with p130 KO (two sgRNAs [#2 and #4]). Two-way ANOVA with Tukey post hoc test. (H and J) Cell-cycle analysis of PAF KD with p130 KO (sgCtrl versus p130 KO) in A549 and H1792 cells; PI staining and FACS. (I and K) Quantification of cell-cycle phases in H and J.
(L–O) PAF per se induces cell quiescence exit and dissociation of p130 from DREAM complex target promoters. (L) Immunoblotting validation of PAF ectopic expression in 3T3 cells, in which PAF is not expressed. (M) PAF ectopic expression reduces the serum starvation-induced G0/G1 phase of 3T3 cells; PI staining-FACS analysis. (N) Quantification of cell-cycle phases. (O) PAF inhibits the recruitment of p130 to the DREAM complex target gene promoters of serum-starved 3T3 cells. ChIP-qPCR analysis of the DREAM target gene promoters (Plk1 and Ccnb1) from each cell line (3T3, 3T3 in serum starvation, and 3T3 ectopically expressing PAF in serum starvation); the ActB promoter served as a negative control.
Representative images are shown. Error bars indicate SEM. ∗p < 0.05 and ∗∗p < 0.01.