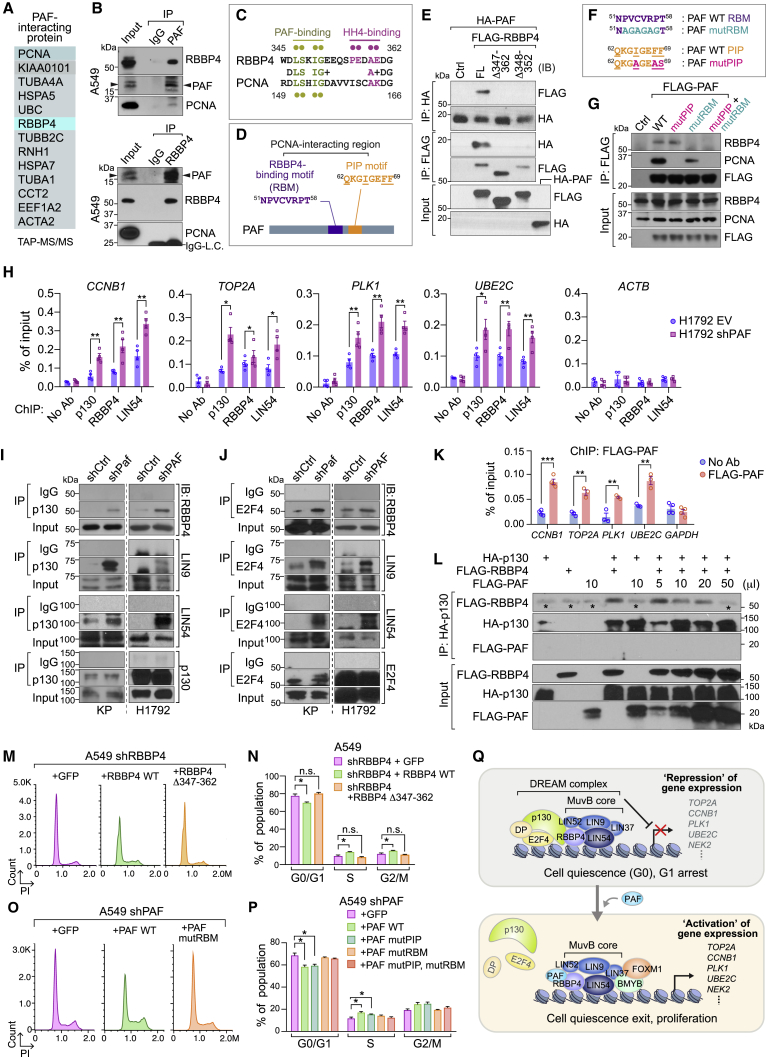
Figure 6.
PAF binds to RBBP4 and antagonizes DREAM complex formation by inhibiting p130 recruitment
(A) Identification of PAF-interacting proteins. HeLa S3 cells stably expressing PAF were processed for TAP-MS/MS. PAF-interacting protein candidates were listed.
(B) Endogenous interaction between PAF and RBBP4. Co-immunoprecipitation (IP)-immunoblotting (IB) analysis in A549 cells. PAF-IP (upper panel) and RBBP4-IP (lower panel). Immunoglobulin IgG, negative control for IP; LC, light-chain of IgG.
(C) Comparative amino acid sequence analysis of potential PAF binding motif in RBBP4 with that in PCNA. Pale green dots, AAs for PAF binding in RBBP4 and PCNA; violet dots, AAs for histone H4 (HH4) binding in RBBP4.
(D) Potential RBBP4-binding motif (RBM) in PAF. Putative RBM (51NPVCVRPT58) is located in PCNA-interacting region. Of note, PAF binds to PCNA via PIP motif (62QKGIGEFF69) and 51NPVCVRPT58 region (De Biasio et al., 2015).
(E) In vitro binding assay of mutant RBBP4 (Δ347–362 and Δ348–352) with PAF. Each protein generated by in vitro transcription and translation was used for coIP and IB.
(F and G) Putative RBM sequence of PAF is required for RBBP4 binding. Amino acid sequences of PAF wild-type (PAF WT), PAF RBM mutant (PAF mutRBM), and PAF PIP mutant (PAF mutPIP) (F). Binding assay of RBBP4 with FLAG-tagged PAF WT and PAF mutRBM (G). Stably transfected A549 cells were used for coIP and IB.
(H) Enrichment of the DREAM components on target gene promoters. The promoter occupancy of p130, RBBP4, and LIN54 on the representative DREAM complex target gene promoters (CCNB1, TOP2A, PLK1, and UBE2C) were analyzed using ChIP-qPCR using human (H1792; control: EV [empty vector] versus shPAF).
(I and J) Increased association of p130-E2F4 with MuvB complex by PAF depletion. CoIP analysis of DREAM complex using anti-p130 (I) or anti-E2F4 (J) antibodies in control versus PAF-KD KP and H1792 cells. IgG served as a negative control for coIP.
(K) Enrichment of PAF on the DREAM target gene promoters. A549 cells stably expressing FLAG-PAF were analyzed using ChIP-qPCR. No-antibody condition and GAPDH served as negative controls.
(L) Inhibition of p130-RBBP4 binding by PAF. In vitro competition assay of p130, RBBP4, and PAF. In vitro transcribed and translated p130, RBBP4, and PAF proteins were mixed and processed for coIP and IB. Asterisks indicate the IgG heavy chain.
(M and N) The replacement of RBBP4 with RBBP4 Δ347–362 mutant induced G0/G1 arrest. The cell-cycle phases were analyzed in A549 cells that transiently expressed shRBBP4 with GFP (control), RBBP4 wild-type (RBBP4 WT), or RBBP4 Δ347–362 mutant (RBBP4 Δ347–362). PI staining with FACS analysis (M) and the quantification of cell-cycle phases (N).
(O and P) The PAF mutRBM could not rescue the G0/G1 arrest induced by PAF depletion. The cell-cycle phases were analyzed in A549 shPAF cells that stably expressed GFP (control), PAF wild-type (PAF WT), or PAF mutRBM. PI staining with FACS analysis (O) and the quantification of cell-cycle phases (P).
(Q) The illustration of the working model, PAF-deregulated DREAM complex for cell quiescence exit and proliferation. PAF dissociates p130-E2F4, gene repression unit, from the DREAM complex, leading to transactivation of the cell proliferative genes (DREAM targets) and promote cell quiescence exit and proliferation of lung cancer cells.
Representative images are shown. Error bars indicate SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.