Figure 3: Structural analysis of titin UN2A-Ig81.
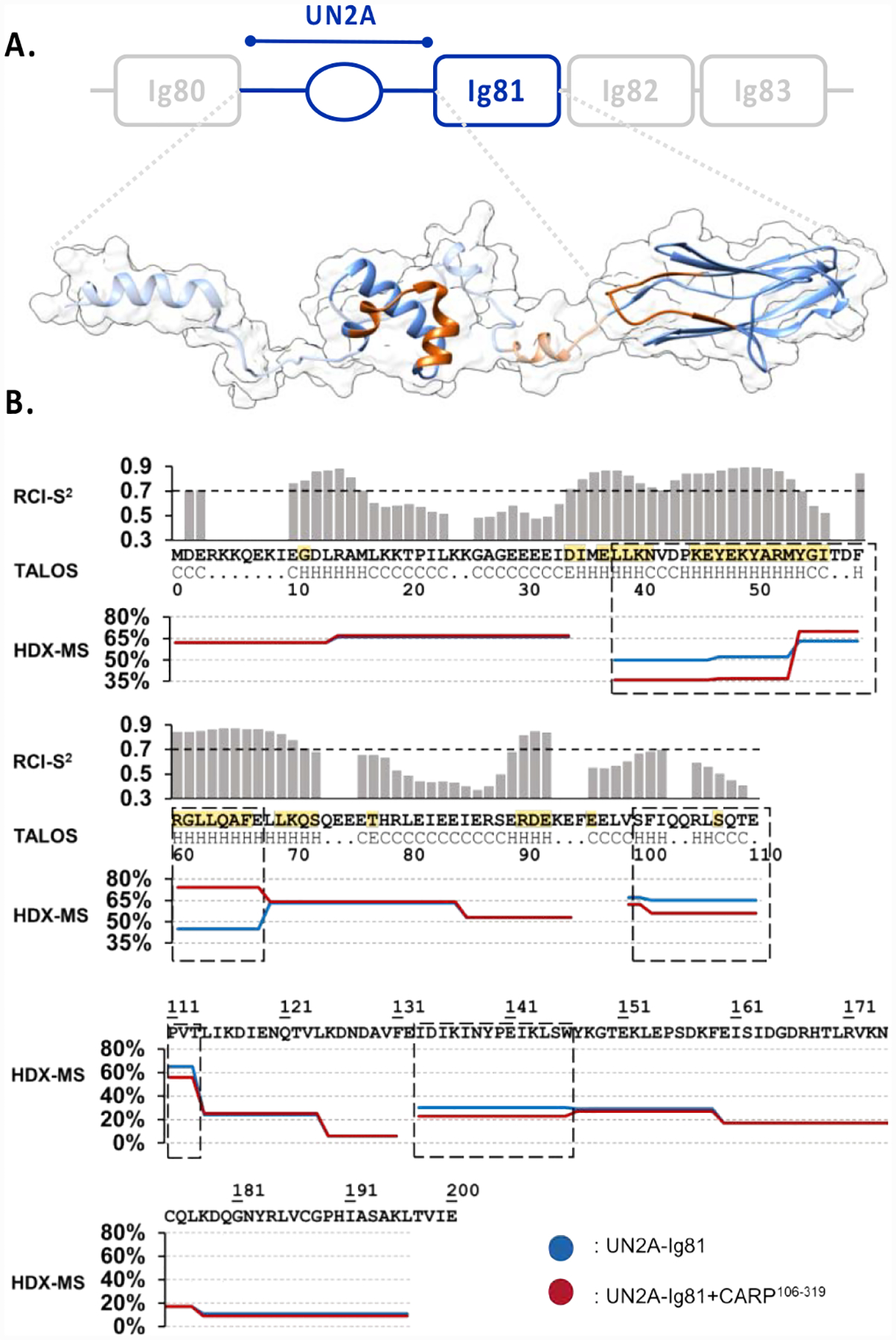
A. Proposed 3D-model of UN2A-Ig81. The experimentally determined, structured domains (UN2A34−73 and Ig81) are shown as solid ribbons, where UN2A34−73 corresponds to the top NMR-guided CS-ROSETTA model and Ig81 is PDB entry 5JOE. Predicted segments are displayed as a semi-transparent ribbon based on TALOS-N output. The model shown is a representative conformation among those coexisting in solution as a result of the flexible semi-helical linker segments. No rotational order on the orientation of domains is implied. The CARP-binding interface identified using HDX-MS is shown in orange; B. NMR-based TALOS-N analysis of UN2A (upper panel). The secondary structure of each residue is below the UN2A sequence (central panel): H indicates α-helix, E is β-strand and C is coil. RCI-S2 values are plotted in histogram format, where a value <0.7 is considered to indicate inherent residue flexibility. Assigned secondary structure and RCI-S2 values indicate the existence of an ordered, helical central fold in the UN2A sequence flanked by flexible N- and C- terminal tails. Residues identified by NMR to be influenced by CARP binding are in yellow. HDX-MS DRFU values for UN2A-Ig81 (lower panel) after 0.5min incubation time for both free (blue) and CARP-bound (red) samples. Sequences boxed (dash line) show significant differences in DRFU values, which is indicative of their participation in CARP binding.
