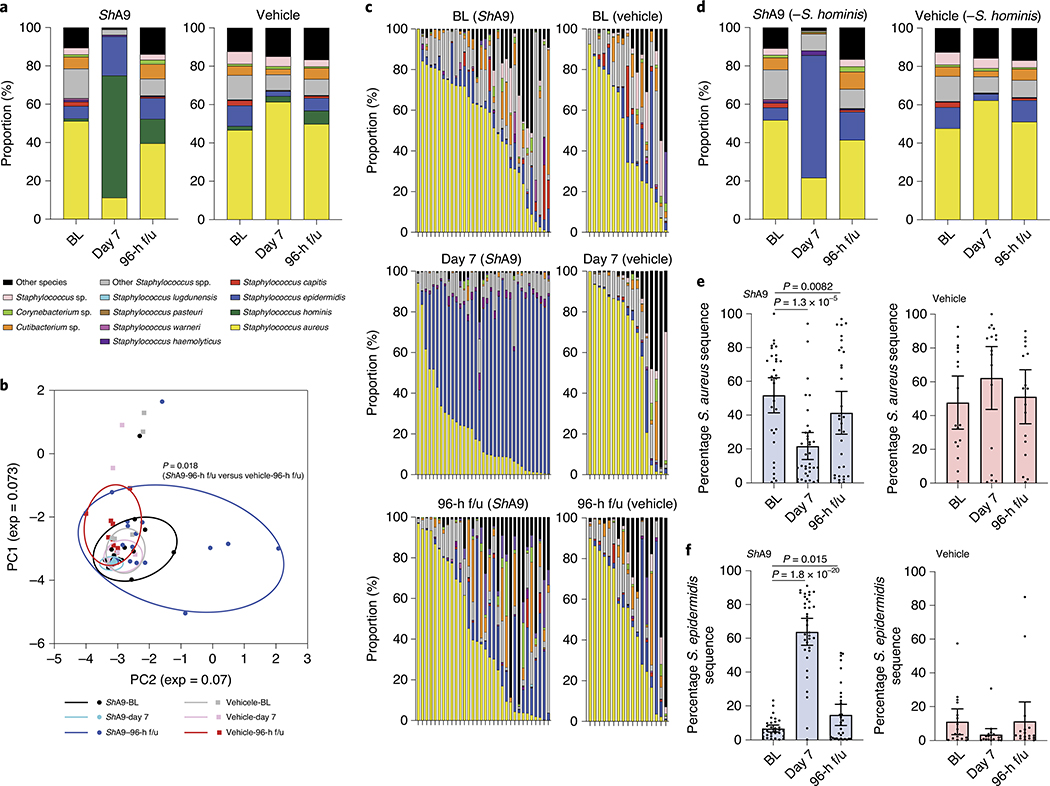
Fig. 5 |. Bacterial species on the skin of participants treated with ShA9.
a, Full-length 16S sequencing analysis of the relative abundance of bacterial species on lesional skin of AD participants at baseline (BL), after 1-week treatment with ShA9 or vehicle (day 7) and 96 h after the end of treatment (96-h f/u). b, PCA plot displaying composition of bacterial community at BL, day 7 and 96-h f/u. Statistical analyses were conducted by paired, parametric Student’s t-test on the relative distance of each dot. c, Relative proportion of bacterial species on individuals treated by ShA9 or vehicle is shown after subtraction of S. hominis. d, Individual subject data from c are averaged showing bacterial species on lesional skin at BL, day 7 and 96-h f/u. e,f, Relative abundance of S. aureus 16S rRNA gene (e) and S. epidermidis 16S rRNA gene (f) determined from c. Data represent average (a and d) or mean ± 95% CI (e and f) (ShA9, BL: n = 33; ShA9, day 7: n = 33; ShA9, 96-h f/u: n = 33; vehicle, BL: n = 16; vehicle, day 7: n = 17; vehicle, 96-h f/u: n = 17 independent subjects) (a–f). Only data from >1,000 contigs were shown and data with <1,000 contigs were excluded (a–f). The P value was calculated using the two-tailed, paired, parametric Student’s t-test (b, e and f).