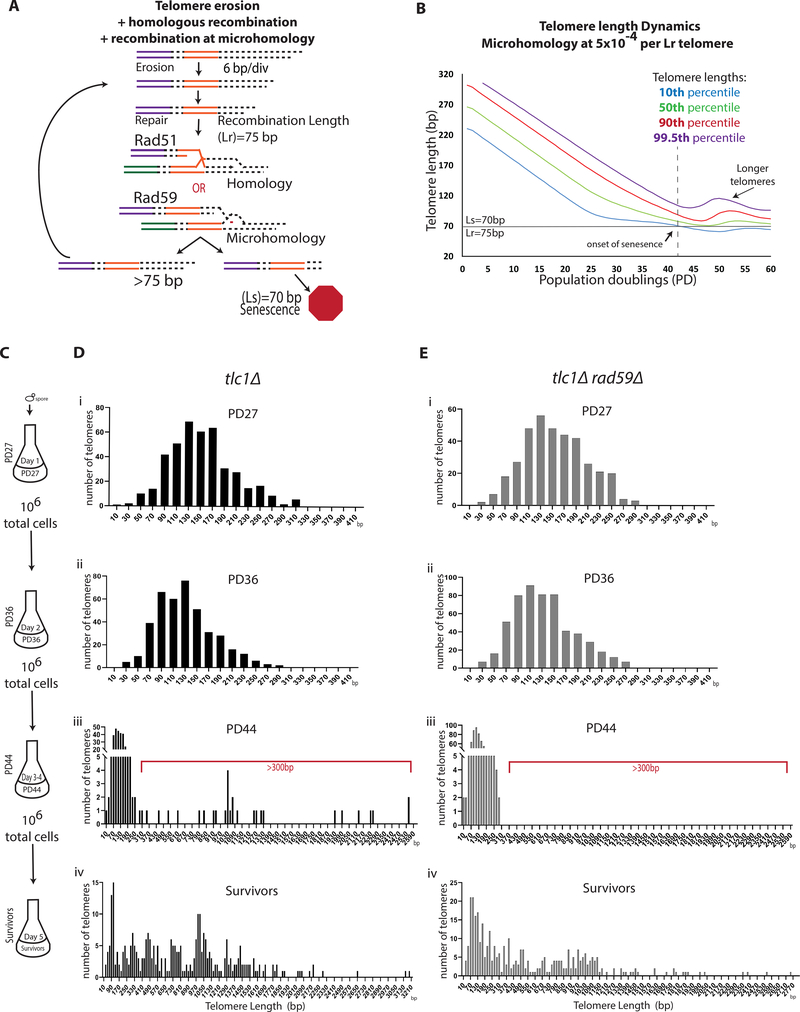
Figure 4. Rad59-mediated recombination mediates the transition from ALT precursors to survivors.
(A-B) Computational modeling of telomere erosion in the presence of recombination at homology and microhomology (see STAR Methods).
(A) Schematic.
(B) The dynamics of the entire distribution of the telomere lengths with parameters (erosion rate=6bp/div., Ls=70, and Lr=75) and with recombination at microhomology at frequency of 5×10−4.
(C) Cell passaging scheme for PacBio sequencing analysis.
(D) Analysis of telomere lengths by PacBio sequencing in tlc1Δ cells passaged, as shown in (C). Telomere length distribution is shown in 20bp bins indicated by the middle value of the bin. Red bracket indicates telomere lengths compatible with ALT survivors. (See also Figure S3)
(E) Analysis of tlc1Δrad59Δ cells like (D).
(See also Figures S3 and S4)