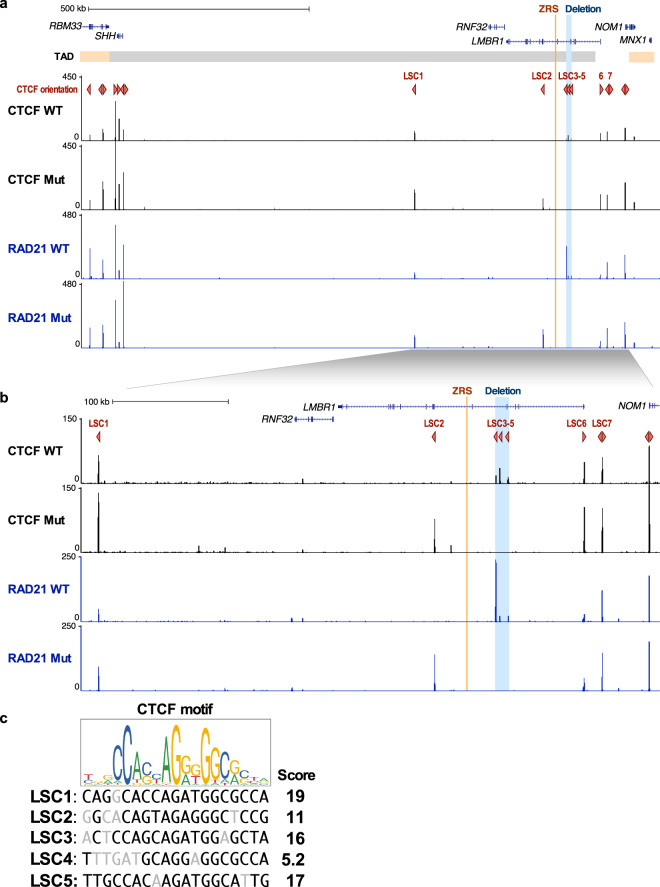
Fig. 3. CTCF and RAD21 distribution in the LMBR1-SHH locus.
a CTCF and RAD21 ChIP-seq enrichment in lymphoblastoid cells from wildtype (WT) and proband (Mut) at the LMBR1-SHH locus. GM12878 (lymphoblastoid cell line) TAD boundaries are shown in orange and gray horizontal bar. ZRS and the acheiropodia-associated deleted region are shown in orange and blue vertical lines respectively. CTCF orientations are shown as red triangles. The Y-axis is the signal P-value to reject the null hypothesis that the signal at that location is present in the control. b Zoom in of the region around the LMBR1 gene. c CTCF motif from JASPAR63 [http://jaspar.genereg.net/] and CTCF motif scores, as assigned by FIMO33, overlapping CTCF peaks in the LMBR1 locus.